The human gastrointestinal tract is a bustling ecosystem, inhabited by trillions of microbial inhabitants that are indispensable not only for digestion but also for shaping the immune landscape of their host. These microbial communities establish a complex and dynamic dialogue with the host’s immune system, mediating a fine balance between tolerance and defense. The molecular mechanisms by which the host selectively identifies and modulates specific bacterial members amidst this vast microbial diversity have remained largely elusive—until now.
In a groundbreaking study published in Nature on May 14, 2025, researchers from the Shanghai Institute of Nutrition and Health and the Center for Excellence in Molecular Cell Science at the Chinese Academy of Sciences unveiled a novel molecular pathway that facilitates intimate crosstalk between the intestinal epithelium and commensal bacteria. The research, spearheaded by Professors QIAN Youcun and SONG Xinyang, sheds light on how apolipoprotein L family members, particularly APOL9, recognize distinct bacterial lipid signatures to orchestrate mucosal immune responses.
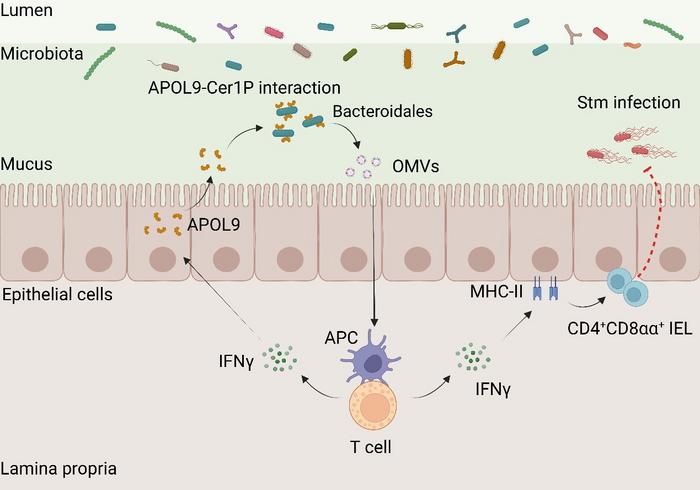
The investigation commenced with a comparative proteomic analysis contrasting the gut epithelium of germ-free mice—lacking any microbiota—with that of conventional pathogen-free mice harboring complex microbial consortia. This approach led to the striking discovery that APOL9 is expressed at markedly higher levels within the intestinal lining of colonized animals, suggesting a microbe-dependent induction of this protein. Notably, APOL9’s expression is localized predominantly to epithelial cells, positioning it as a prime candidate for mediating host-microbe interactions at the mucosal interface.
Moving beyond correlation, the team engineered an innovative technique termed “APOL9-seq,” which integrates flow cytometric sorting with high-throughput sequencing to map the bacterial species targeted by APOL9. Unexpectedly, APOL9 exhibited high-affinity binding specifically to members of the order Bacteroidales, a dominant resident group within the mammalian gut microbiota. This specificity hints at a refined host capacity to discriminate among microbial neighbors based on molecular cues.
Delving deeper, the molecular underpinnings of this selective recognition were deciphered. The researchers uncovered that APOL9 binds to a unique sphingolipid derivative on the bacterial surface, ceramide-1-phosphate (Cer1P). Genetic ablation of bacterial Cer1P biosynthesis abolished APOL9 binding, conclusively proving that Cer1P serves as the bacterial “molecular fingerprint” recognized by this host protein. This represents the first known instance of a host protein directly engaging a bacterial lipid moiety to modulate immune crosstalk.
Importantly, APOL9 does not exert bactericidal effects; instead, it induces Bacteroidales to release outer membrane vesicles (OMVs)—nano-scaled lipid bilayer-enclosed spheres that package numerous bacterial molecules. The secretion of OMVs represents a subtle mechanism of communication, delivering microbial signals to host cells while sparing the bacteria themselves. These OMVs were experimentally demonstrated to potentiate the host interferon-gamma (IFN-γ) signaling pathway and augment the expression of major histocompatibility complex class II (MHC-II) molecules on intestinal epithelial cells.
The upregulation of MHC-II molecules by OMVs is particularly significant because it intensifies antigen presentation to CD4⁺CD8αα⁺ intraepithelial T cells—a specialized subset critical for maintaining intestinal immune homeostasis. This cellular circuit appears to form a feedback loop whereby APOL9-mediated recognition of Bacteroidales enhances mucosal immune readiness without triggering overt inflammation or microbial eradication, thereby preserving the delicate symbiotic relationship.
To interrogate the physiological relevance of APOL9, in vivo mouse models deficient in the APOL9 gene were employed. These knockout mice exhibited increased susceptibility to Salmonella infection, accompanied by attenuated IFN-γ responses and elevated systemic bacterial dissemination. Intriguingly, administration of Bacteroidales-derived OMVs to these mice restored immune function and mitigated infection severity, underscoring the therapeutic potential of harnessing OMV-mediated pathways.
Professor QIAN Youcun remarked on the evolutionary significance of the findings: “The exquisitely specific interaction between APOL9 and bacterial Cer1P embodies a molecular dialogue sculpted by millennia of host-microbiota coevolution. Our work opens avenues to explore analogous mechanisms in humans, particularly involving the homologous apolipoprotein L2, and raises the prospect of enhancing intestinal immunity by modulating these interactions.”
This pioneering study overturns traditional views that host defense proteins primarily exert antimicrobial functions by killing pathogens. Instead, it introduces a paradigm wherein host proteins selectively engage bacterial lipids to elicit beneficial immune modulatory effects, exemplifying a sophisticated mutualistic communication channel. Such discoveries illuminate the intricate language spoken between host and microbiome and propel forward the quest to develop targeted therapeutics that reinforce the intestinal barrier through microbiota-directed immunomodulation.
The implications extend beyond fundamental immunology into clinical realms, as dysregulation of host-microbial interactions is implicated in a variety of inflammatory bowel diseases and systemic immune disorders. Manipulating APOL9/APOL2 pathways or harnessing OMVs could therefore inspire novel strategies to restore mucosal integrity and immune equilibrium in disease contexts.
In sum, this study establishes a new molecular axis through which the host senses specific microbial lipids to orchestrate localized immune education and tolerance. By revealing how apolipoproteins selectively target bacterial Cer1P to stimulate outer membrane vesicle release and downstream IFN-γ-MHC-II signaling, it broadens our understanding of immune-microbiota symbiosis and sets the stage for innovative interventions aimed at fortifying gut health.
Subject of Research: Host-microbe interactions in the gut; molecular recognition of bacterial lipids by apolipoproteins; mucosal immunity modulation
Article Title: Targeting symbionts by apolipoprotein L proteins modulates gut immunity
News Publication Date: 14-May-2025
Web References:
10.1038/s41586-025-08990-4
Image Credits: Image by Prof. QIAN’s group
Keywords: Intestines, Gut microbiota, Immune modulation, Apolipoprotein L9, Outer membrane vesicles, Ceramide-1-phosphate, Interferon-gamma, MHC class II, Bacteroidales, Mucosal immunity, Host-microbe interaction
Tags: apolipoprotein L familycommensal bacteria recognitioncrosstalk between gut epithelium and bacteriagastrointestinal microbiota researchgut microbiome communicationhost-microbe interactionsimmune system modulationlipid-based signaling pathwaysmicrobial diversity in humansmucosal immune responsesproteomic analysis in miceShanghai Institute of Nutrition and Health