Lawrence Livermore researchers in conjunction with collaborators at University of California (link is external), Los Angeles have found that some cells build intracellular compartments that allow the cell to store metals and maintain equilibrium.
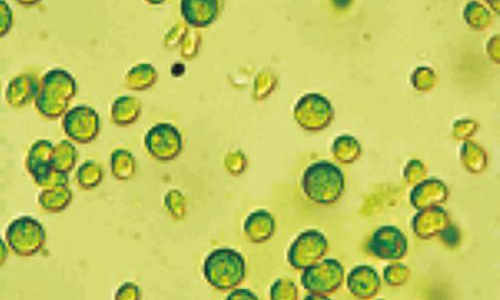
Lawrence Livermore researchers have discovered that cells of the alga Chlamydomonas Reinhardti (seen under a microscope) build a “pantry” to store metal and maintain equilibrium. Nearly 40 percent of all cells in all organisms require metal ions such as zinc, copper, manganese or iron for activity
Nearly 40 percent of all proteins require metal ions such as zinc, copper, manganese or iron for activity.
“We don’t understand very well how cells maintain balance when the cell is stressed by metal excess or metal deficiency,” said LLNL researcher Jennifer Pett-Ridge. “By storing the metal in a special intracellular compartment, the cell creates a bit of a pantry cupboard for itself and can better maintain its equilibrium.”
How this matchmaking of metals and proteins occurs with such precision has been a puzzle in cell physiology. The researchers shed new light into how such partnerships are maintained even under stressful conditions of metal deprivation.
The research team studied the copper content in Chlamydomonas reinhardtii, a single-cell green alga. The copper quota in this cell is maintained at a relatively constant level over orders of magnitude of extracellular copper. However, copper will hyper-accumulate when Chlamydomonas is starved for zinc.
In their preliminary work, the researchers at UCLA measured metal concentrations in bulk—large numbers of cells were digested and analyzed on a mass spectrometer— and showed intriguing patterns; copper concentrations tended to increase when cells were grown without zinc.
To understand this phenomenon at a single cell scale, “we needed high resolution elemental imaging to understand how the cell builds the pantry cupboard,” Pett-Ridge said. “Our work looking at spatial distribution shows the mechanism of how the cell achieves metal storage—they put the copper into a little vacuole type container.” (A vacuole is an enclosed compartment, which is filled with water containing inorganic and organic molecules including enzymes).
By using the combined power of fluorescent metal sensors, transmission electron microscopy and secondary ion mass spectrometry (NanoSIMS—housed at Lawrence Livermore) imaging, the team found that the bulk of copper in zinc-deficient cells is concentrated in electron-dense and phosphate- and calcium-rich bodies (the so-called pantry cupboard). This imprisoned form of the metal is hidden from cellular sensors of copper. But this trapping of copper is not a dead end for the metal; when the zinc starvation stress is alleviated, copper is released and is used preferentially over extracellular sources of copper for biosynthesis.
“Our research shows the great lengths cells will take to restrict adverse reactions with copper, even at the expense of limiting cell activity,” said UCLA postdoc Marcus Miethke, co-first author of the paper appearing in the journal Nature Chemical Biology.
Both lead authors, A. Hong-Hermesdorf and Miethke, were supported by a German DAAD Research fellowship to work on metal deficiency stress specifically in Chlamydomonas. As part of a DOE Office of Biological and Environmental Research-Genomic Sciences Biofuels “SFA” (Scientific Focus Area), LLNL has been working to image the subcellular distribution of metals in microbes for the past 4-5 years. The primary focus of this work has been collaboration with Sabeeha Merchant’s group (UCLA) to image the subcellular distribution of copper and zinc in Chlamydomonas.
The high-resolution copper imaging was made possible by LLNL’s recently installed Hyperion II ion source. This is currently a one-of-a-kind capability that allows researchers to image metals with as good as 50-nanometer lateral resolution. “With the Hyperion, we can image trace metals with unprecedented resolution,” said LLNL researcher Peter Weber. The Livermore team is using this capability in ongoing studies of Chlamydomonas in partnership with Merchant’s Lab.
Story Source:
The above story is based on materials provided by Lawrence Livermore National Laboratory.