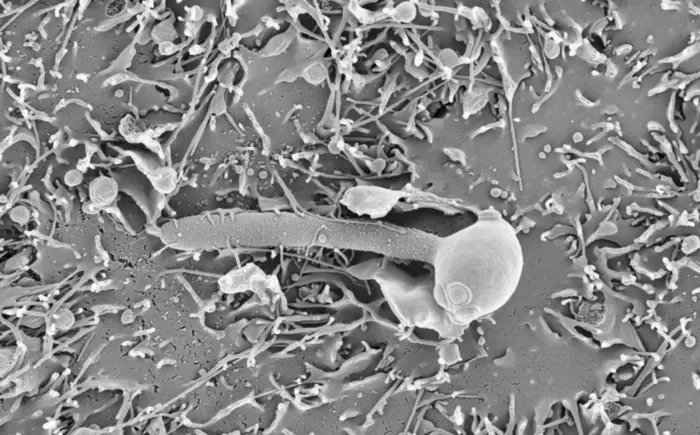
Barcelona, 12 January 2024 – Global fungal infections, which affect one billion people and cause 1.5 million deaths each year, are on the rise due to the increasing number of medical treatments that heighten vulnerability. Patients undergoing chemotherapy or immunosuppressive treatments after organ transplant often present compromised immune systems. Given the emergence of resistant strains, the limited variety of current antifungal drugs as well as their cost and side effects, the treatment of these infections is challenging and brings about an urgent need for more effective treatments.
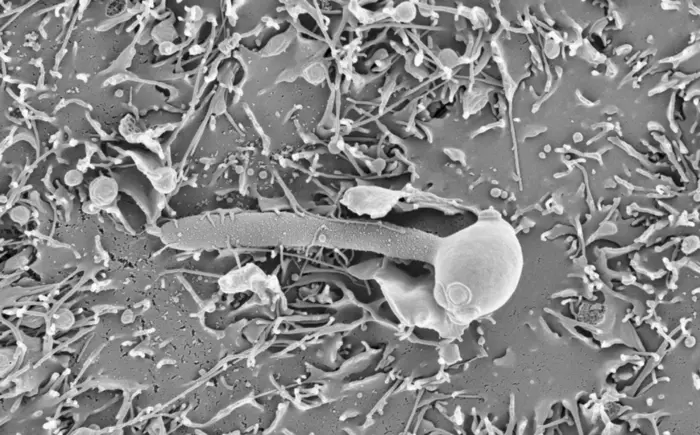
Credit: IRB Barcelona
Barcelona, 12 January 2024 – Global fungal infections, which affect one billion people and cause 1.5 million deaths each year, are on the rise due to the increasing number of medical treatments that heighten vulnerability. Patients undergoing chemotherapy or immunosuppressive treatments after organ transplant often present compromised immune systems. Given the emergence of resistant strains, the limited variety of current antifungal drugs as well as their cost and side effects, the treatment of these infections is challenging and brings about an urgent need for more effective treatments.
In this context, a team from the Institute for Research in Biomedicine (IRB Barcelona) and the Barcelona Supercomputing Center – Centro Nacional de Supercomputación (BSC-CNS), led by the ICREA researcher Dr. Toni Gabaldón, has identified hundreds of genes subject to recent, clinically-relevant selection in six species of the fungal pathogen Candida.
“This work highlights how these pathogens adapted to humans and antifungal drugs and provides valuable knowledge that could lead to better treatments for Candida infections,” explains Dr. Gabaldón, head of the Comparative Genomics lab at IRB Barcelona and the BSC.
More than 2,000 genomes from 6 different species
The study delves into the evolutionary landscape of Candida pathogens by analysing approximately 2,000 genomes from clinical samples of six major Candida species. These genomes are stored in public databases. The researchers compared these genomes to a reference, creating a comprehensive catalogue of genetic variants.
Building on previous work addressing drug-resistant strains, the researchers conducted a Genome-Wide Association Study (GWAS) to identify genetic variants linked to antifungal drug resistance in clinical isolates. This approach provided insights into both known and novel mechanisms of resistance towards seven antifungal drugs in three Candida species. “Additionally, a concerning finding has arisen from the study: the potential spread of resistance through mating between susceptible and resistant strains, contributing to the prevalence of drug-resistant Candida pathogens,” explains Dr. Miquel Àngel Schikora-Tamarit, a postdoctoral researcher in the same lab and first author of the study.
In addition, by focusing on variants acquired recently among clinical strains, the researches detected shared and species-specific genetic signatures of recent selection that inform on which adaptations might be needed to thrive and spread in human-related environments.
Beyond the novel insights into the adaptation of Candida, the study provides a valuable resource, namely a comprehensive catalogue of variants, selection signatures, and drivers of drug resistance. This knowledge not only contributes to our understanding of these infections but also lays the groundwork for future experiments and potential advancements in the development of more effective treatments for Candida infections.
This work was funded by the Spanish Ministry of Science, Innovation and Universities, The European Research Council, and “la Caixa” Foundation.
Journal
Nature Microbiology
DOI
10.1038/s41564-023-01547-z