Many biological structures of impressive beauty and sophistication arise through processes of self-assembly. Indeed, the natural world is teeming with intricate and useful forms that come together from many constituent parts, taking advantage of the built-in features of molecules.
Credit: Petr Sulc lab
Many biological structures of impressive beauty and sophistication arise through processes of self-assembly. Indeed, the natural world is teeming with intricate and useful forms that come together from many constituent parts, taking advantage of the built-in features of molecules.
Scientists hope to gain a better understanding of how this process unfolds and how such bottom-up construction can be used to advance technologies in computer science, materials science, medical diagnostics and other areas.
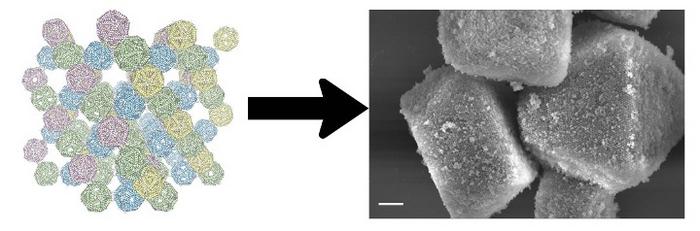
In new research, Arizona State University Assistant Professor Petr Sulc and his colleagues have taken a step closer to replicating nature’s processes of self-assembly. Their study describes the synthetic construction of a tiny, self-assembled crystal known as a “pyrochlore,” which bears unique optical properties.
The key to creating the crystal is the development of a new simulation method that can predict and guide the self-assembly process, avoiding unwanted structures and ensuring the molecules come together in just the right arrangement.
The advance provides a steppingstone to the eventual construction of sophisticated, self-assembling devices at the nanoscale — roughly the size of a single virus.
The new methods were used to engineer the pyrochlore nanocrystal, a special type of lattice that could eventually function as an optical metamaterial, “a special type of material that only transmits certain wavelengths of light,” Sulc says. “Such materials can then be used to produce so-called optical computers and more sensitive detectors, for a range of applications.”
Sulc is a researcher in the Biodesign Center for Molecular Design and Biomimetics, the School of Molecular Sciences and the Center for Biological Physics at Arizona State University.
The research appears in the current issue of the journal Science.
From chaos to complexity
Imagine placing a disassembled watch into a box, which you then shake vigorously for several minutes. When you open the box, you find an assembled, fully functional watch inside. Intuitively, we know that such an event is nearly impossible, as watches, like all other devices we manufacture, must be assembled progressively, with each component placed in its specific location by a person or a robotic assembly line.
Biological systems, such as bacteria, living cells or viruses, can construct highly ingenious nanostructures and nanomachines — complexes of biomolecules, like the protective shell of a virus or bacterial flagella that function similarly to a ship’s propeller, helping bacteria move forward.
These and countless other natural forms, comparable in size to a few dozen nanometers — one nanometer is equal to one-billionth of a meter, or roughly the length your fingernail grows in one second — arise through self-assembly. Such structures are formed from individual building blocks (biomolecules, such as proteins) that move chaotically and randomly within the cell, constantly colliding with water and other molecules, like the watch components in the box you vigorously shake.
Despite the apparent chaos, evolution has found a way to bring order to the unruly process.
Molecules interact in specific ways that lead them to fit together in just the right manner, creating functional nanostructures inside or on the cell’s surface. These include various intricate complexes inside cells, such as machinary that can replicate entire genetic material. Less intricate examples, but quite complex nevertheless, include self-assembly of the tough outer shells of viruses, whose assembly process Sulc also previously studied with his colleague, Banu Ozkan from ASU’s Department of Physics.
Crafting with DNA
For several decades, the field of bionanotechnology has worked to craft tiny structures in the lab, replicating the natural assembly process seen in living organisms. The technique generally involves mixing molecular components in water, gradually cooling them and hoping that when the solution reaches room temperature, all the pieces will fit together correctly.
One of the most successful strategies, known as DNA bionanotechnology, uses artificially synthesized DNA as the basic building block. This molecule of life is not only capable of storing vast troves of genetic information — strands of DNA can also be designed in the lab to connect with each other in such a way that a clever 3D structure is formed.
The resulting nanostructures, known as DNA origami, have a range of promising applications, from diagnostics to therapy, where, for example, they are being tested as a new method of vaccine delivery.
A significant challenge lies in engineering molecule interactions to form only the specific, pre-designed nanostructures. In practice, unexpected structures often result due to the unpredictable nature of particle collisions and interactions. This phenomenon, known as a kinetic trap, is akin to hoping for an assembled watch after shaking a box of its parts, only to find a jumbled heap instead.
Maintaining order
To attempt to overcome kinetic traps and ensure the proper structure self-assembles from the DNA fragments, the researchers developed new statistical methods that can simulate the self-assembly process of nanostructures.
The challenges for achieving useful simulations of such enormously complex processes are formidable. During the assembly phase, the chaotic dance of molecules can last several minutes to hours before the target nanostructure is formed, but the most powerful simulations in the world can only simulate a few milliseconds at most.
“Therefore, we developed a whole new range of models that can simulate DNA nanostructures with different levels of precision,” Sulc says. “Instead of simulating individual atoms, as is common in protein simulations, for example, we represent 12,000 DNA bases as one complex particle.”
This approach allows researchers to pinpoint problematic kinetic traps by combining computer simulations with different degrees of accuracy. Using their optimization method, researchers can fine-tune the blizzard of molecular interactions, compelling the components to assemble correctly into the intended structure.
The computational framework established in this research will guide the creation of more complex materials and the development of nanodevices with intricate functions, with potential uses in both diagnostics and treatment.
The research work was carried out in collaboration with researchers from Sapienza University of Rome, Ca’ Foscari University of Venice and Columbia University in New York.
Journal
Science
DOI
10.1126/science.adl5549
Method of Research
Experimental study
Subject of Research
Not applicable
Article Title
Inverse design of a pyrochlore lattice of DNA origami through model-driven experiments
Article Publication Date
16-May-2024




