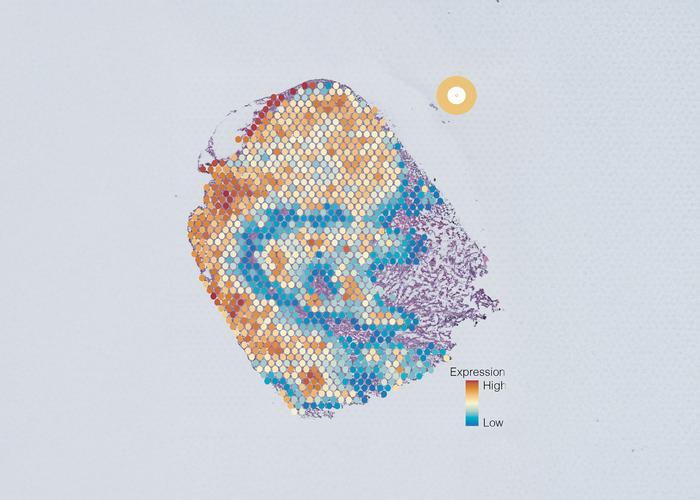
A groundbreaking multinational study, co-led by the Garvan Institute of Medical Research, has unveiled a pioneering artificial intelligence tool designed to dissect and characterize the cellular diversity within tumors with unprecedented precision. This novel approach promises to revolutionize the landscape of cancer treatment, particularly for historically challenging forms like triple-negative breast cancer, by enabling personalized therapies that specifically target all distinct cancer cell types harbored within a single tumor mass.
Cancer is notoriously heterogeneous. Tumors are not homogenous masses but intricate ecosystems composed of varied populations of cells, each exhibiting unique genetic and behavioral traits. This intratumoral heterogeneity is a formidable obstacle in oncology, often underpinning variable responses to treatment and disease relapse. Traditional cancer therapies typically assume uniformity, targeting a dominant mechanism shared by most tumor cells. However, residual subpopulations resistant to such interventions can survive and drive cancer recurrence, underscoring the urgent need for more precise cellular characterization tools.
Until now, the scientific community has struggled to systematically classify and understand the nuanced differences between individual cancer cells residing side-by-side in the tumor microenvironment. The subtle molecular and functional distinctions among these cells have remained elusive, creating a gap between biological complexity and therapeutic strategy. Understanding how to deconvolute this complexity is critical for devising treatments that can comprehensively eliminate all malignant cell types within a tumor.
.adsslot_fMH4nt5Jeu{width:728px !important;height:90px !important;}
@media(max-width:1199px){ .adsslot_fMH4nt5Jeu{width:468px !important;height:60px !important;}
}
@media(max-width:767px){ .adsslot_fMH4nt5Jeu{width:320px !important;height:50px !important;}
}
ADVERTISEMENT
Addressing this knowledge gap, the research coalition developed an advanced AI-driven framework termed AAnet, an artificial neural network specifically tailored to analyze single-cell gene expression data. Unlike conventional clustering methods that often oversimplify cellular phenotypes, AAnet employs innovative deep learning algorithms to detect continuous patterns in gene expression, thus capturing the high-dimensional diversity of tumor cells with remarkable granularity.
Applying AAnet to extensive datasets derived from murine models of triple-negative breast cancer as well as human tissue samples representing ER-positive, HER2-positive, and triple-negative subtypes, the researchers identified five distinct cancer cell archetypes coexisting within tumors. Each archetype exhibits unique gene expression signatures that correspond to deeply divergent biological functions and behaviors, including differential proclivities for proliferation, metastasis, and resistance, which collectively influence patient prognosis.
These five archetypes effectively distill the complex spectrum of cancer cell states into discrete, biologically meaningful categories. For instance, some archetypes are enriched for pathways associated with aggressive growth and invasiveness, whereas others exhibit signatures tied to quiescence or metabolic adaption. This classification framework not only simplifies the intricate cellular landscape but also opens a window into understanding spatial tumor architecture and metabolic variation with potential implications for biomarker discovery.
One of the most transformative aspects of this research lies in its potential clinical application. With AAnet, oncologists could move beyond traditional organ-centric and molecular subtyping toward a refined, cell-based classification paradigm. This is pivotal because it recognizes that effective cancer therapy must address the heterogeneity within tumors, designing customized combination regimens that target each cellular archetype’s specific molecular vulnerabilities, thereby maximizing therapeutic efficacy.
The implications are particularly striking for triple-negative breast cancer patients, a subgroup notoriously lacking targeted treatments due to its heterogeneity and aggressiveness. The ability to identify and track the dynamics of these five cell groups before and after chemotherapy offers unprecedented opportunities to tailor interventions dynamically, monitor treatment response in real time, and potentially prevent relapse through early detection of resistant archetypes.
Technologically, AAnet represents a significant leap forward in single-cell analytical methodologies. By leveraging deep learning techniques traditionally used in artificial intelligence research, the model transcends the limitations of earlier clustering algorithms, enabling researchers to map continuous trajectories of cellular states rather than forcing discrete, and sometimes artificial, classifications. This innovation in modeling allows for a more authentic representation of cellular plasticity and diversity within tumors.
Beyond breast cancer, the methodological framework embodied by AAnet holds promise for broader biomedical applications. Its capacity to resolve complex mixtures of cell states could be applicable not only to a variety of cancers but also to other diseases characterized by cellular heterogeneity, such as autoimmune disorders. This platform effectively bridges cutting-edge computational science with translational biology, heralding a new era of personalized medicine driven by AI-powered insights.
In summary, this study exemplifies the profound impact of integrating artificial intelligence with molecular oncology. By systematically unveiling the cellular diversity that underpins tumor behavior, it establishes a powerful foundation for developing next-generation, multi-targeted therapeutic strategies. Continued exploration and validation of these findings could redefine standard-of-care protocols and significantly enhance patient outcomes in oncology.
The team’s work, recently published in the American Association for Cancer Research’s journal Cancer Discovery, reflects collaboration among leaders in computational biology, cancer research, and clinical medicine, illustrating the multidisciplinary nature of modern scientific breakthroughs. It also highlights the synergy between technological innovation and biological inquiry as an essential driver for unraveling complex diseases.
Looking ahead, the researchers aim to extend their analyses longitudinally to monitor how these five archetypes evolve over time and in response to various treatments, providing insights into tumor evolution and mechanisms of resistance. This longitudinal perspective could help optimize timing and combinations of therapies, ultimately realizing the promise of precision oncology in clinical settings.
Subject of Research: Animals
Article Title: AAnet resolves a continuum of spatially-localized cell states to unveil intratumoral heterogeneity
News Publication Date: 24-Jun-2025
Web References: https://doi.org/10.1158/2159-8290.CD-24-0684
Image Credits: Garvan Institute
Keywords: Breast cancer cells, Cancer cells, Cancer, Tumor cells, Neoplastic cells, Cell proliferation, Cell biology, Xenografts, Metastasis, Breast cancer, Metabolic networks, Metabolic pathways, Artificial intelligence, Artificial neural networks, Modeling, Computer modeling
Tags: AI in cancer treatmentartificial intelligence in oncologycancer cell characterization toolsgroundbreaking cancer study resultsintratumoral heterogeneity challengesmultidisciplinary cancer researchnovel approaches to tumor microenvironmentovercoming treatment resistance in cancerpersonalized cancer therapiesprecision medicine for cancertriple-negative breast cancer advancementstumor cellular diversity