An open-source data platform for researchers studying archaea is paving the way for new insights and educational opportunities
Credit: Laboratory of Mecky Pohlschroder
Bioinformatics and big data analyses can reap great rewards for biologists, but it takes a lot of work to generate the datasets necessary to begin. At the same time, researchers around the globe churn out datasets that could be useful to others but are not always widely shared.
To foster scientific exchange and to advance discovery, biologists in the School of Arts & Sciences led by postdoc Stefan Schulze and professor Mecky Pohlschroder have launched the Archaeal Proteome Project (ArcPP), a web-based database to collect and make available datasets to further the work of all scientists interested in archaea, a domain of life composed of microorganisms that can dwell anywhere from deep-sea vents to the human gut.
“This is a very community-focused effort,” says Schulze, who has worked as a postdoc in Pohlschroder’s lab since 2017 and took the lead in developing the ArcPP platform, which is described in a recent Nature Communications paper. “People who are working in different fields and are interested in different biological questions may all be generating proteomics datasets to answer their questions. But those same datasets could be analyzed to answer other questions as well. Our idea was to bring these datasets together in a uniform way to be of use to the whole community.”
Pohlschroder’s lab studies the archaeon Haloferax volcanii as a model organism, a salt-loving species originally isolated from the Dead Sea. While the ArcPP launched with data from only this species, the researchers hope to rapidly grow it to encompass proteomic data–a cataloging of the entire set of proteins contained in an organism–from more archaeal species and even beyond, including other single-celled organisms, such as bacteria.
“The principle of the ArcPP can be seen as similar to the collaboration between medical specialists treating a patient,” Schulze says. “Brain, heart, or kidney specialists all have expert knowledge in their respective fields, but for all of them a blood sample can help to interpret symptoms of a patient. Similar to that blood sample, modern proteomics, which can analyze the whole proteomes of an organism within a single experiment, provide information about various aspects of archaeal cell biology.”
Archaea are a relatively understudied group, but they play important ecological roles, are used for various biotechnological applications, and appear to be the prokaryotic ancestors of eukaryotes. Thus, the field is ripe for novel insights into their biochemistry and function.
Recent advances have made it much simpler to generate the raw data needed to perform proteomics studies with an organism. “Now the bottleneck is how do you effectively analyze it, and what do you make out of this analysis,” Pohlschroder says.
That’s where the ArcPP community comes in. “I might understand why certain proteins are expressed or modified on the cell surface because that’s what we focus on in our lab,” she says, “but our colleagues study other aspects of archaeal biology. By bringing together the community of scientists studying various aspects of archaeal proteomics, ArcPP can provide the research community with an abundance of easily accessible data and also has the expertise and perspectives needed to analyze the data in ways that will yield significant new insights into archaeal biology.”
To develop the ArcPP, the Penn biologists reached out to multiple laboratories around the world to contribute their proteomics datasets for H. volcanii. The data represented analyses of the microbes growing in a broad range of conditions, resulting in a collection that is a massive two terabytes in size.
“We were able to identify 72% of known proteins encoded in the H. volcanii proteome,” Pohlschroder says. “By comparing different culture conditions, we were able to identify proteins that are always present, indicating that they are crucial for cell functions in a variety of environments. Interestingly, for at least 15% of these proteins the function is as of yet completely unknown, highlighting that our understanding of archaeal cell biology is still quite limited.”
Schulze put the platform to the test to see what new information could be gleaned. Together with other members of the group, he used the database to find that, contrary to what was previously believed about H. volcanii, it can express the enzyme urease, which breaks down the nitrogen compound urea, though only in the presence of glycerol as a carbon source. Follow-up experiments at the bench by collaborators from the University of Florida confirmed the finding, offering a proof-of-concept of the power of ArcPP.
“Expressing urease could be important in nitrogen cycling in the environment,” Schulze says, “or even for biotechnology applications.”
Another powerful aspect of ArcPP is its utility for education. Bioinformatics is an invaluable skill for up-and-coming biologists, and analyses that can be done at the computer rather than the lab are a useful way to safely continue scientific discovery amid the COVID-19 pandemic. It’s something that even the high school students that Pohlschroder invites into her lab through the program Penn LENS, short for Laboratory Experience in Natural Sciences, can experience in a hands-on format.
“What I think is fascinating about this project is that you can work with Haloferax volcanii, which is non-pathogenic and fairly easy to work with,” Pohlschroder says, “and pair it with cutting-edge technology but do it in such a way that high school students are capable of making absolutely novel discoveries. It’s something we are definitely thinking about using for the upcoming semester for undergraduates as well since they may not be able to come into the laboratory right away.”
A new study Schulze, Pohlschroder, doctoral student Heather Schiller, and colleagues,
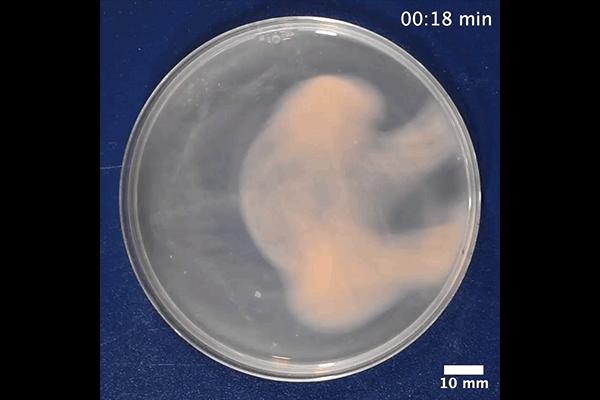
released as a pre-print to bioRxiv, prior to peer-review, also offers hints at how ArcPP might play a role in extending bench-based scientific discoveries. Many archaea, like bacteria, can form biofilms, which are microbial communities of adherent cells embedded in an extracellular polymeric matrix. H. volcanii can form biofilms either on solid surfaces or in liquid media. Schulze had noticed that, when H. volcanii forms a biofilm in liquid media contained in a petri dish, it can rapidly develop an intricate honeycomb pattern upon the removal of the petri dish lid.
After some detective work to find out what is responsible for creating this formation, the researchers ruled out genes responsible for surface adhesion, light, oxygen, humidity, and other variables, and they now believe the driver to be a volatile compound in the air.
While the group is planning further “wet lab” follow-up to determine whether other archaeal species and even certain bacteria do something similar, they also hope to lean on the ArcPP to better understand the mechanism of the honeycomb formation, as the included datasets contain proteomic information from microbes in biofilms as well as growing freely.
“I always see bioinformatics and lab work as a circle,” Schulze says. “You may start with lab work, go to bioinformatics to probe into a finding, but you don’t stop there. You should always go back to the lab–and then back around–to confirm and extend your findings.”
With ArcPP, these biologists are hoping to extend that circle, bringing more researchers–and young people with scientific aspirations–into the fold.
“We are eager to work together with more laboratories in order to extend our analyses to other archaeal species or even bacteria, harvesting the synergistic effects of a broad scientific community,” says Pohlschroder.
###
Mecky Pohlschroder is a professor and undergraduate chair in the Department of Biology in the School of Arts & Sciences at the University of Pennsylvania.
Stefan Schulze is a postdoctoral research in the Pohlschroder lab in the Department of Biology in the School of Arts & Sciences at the University of Pennsylvania.
Pohlschroder and Schulze’s coauthors on the Nature Communications paper are Penn’s Georgio Legerme; the University of Florida’s Zachary Adams and Julie Maupin-Furlow; the National University of Mar del Plata’s Micaela Cerletti, Rosana De Castro, María Inés Giménez, and Roberto A. Paggi; the University of Regensburg’s Sébastien Ferreira-Cerca and Robert Knüppel; the Heidelberg University’s Christian Fufezan; the University of Münster’s Michael Hippler; the Max Planck Institute of Biophysical Chemistry’s Zivojin Jevtic, Christof Lenz, and Henning Urlaub; Ulm University’s Anita Marchfelder; the Max Planck Institute of Biochemistry’s Friedhelm Pfeiffer; and Ruhr University Bochum’s Ansgar Poetsch.
Pohlschroder and Schulze’s coauthors on the bioRxiv pre-print are co-first author Heather Schiller and Zuha Metan of Penn’s Biology Department, and Theopi Rados and Alexandre W. Bisson Filho of Brandeis University.
Support for the research came from the German Research Foundation, National Science Foundation (grants 1817518 and 1642283), U.S. Department of Energy (Grant DE-FG02-05ER15650), National Institutes of Health (Grant GM57498), and National Agency for the Promotion of Science and Technology-ANPCyT and MINCyT-BMBF (Argentina-Germany).
Media Contact
Katherine Unger Baillie
[email protected]
Original Source
https:/
Related Journal Article
http://dx.




