Credit: Frank Schmitges et al.
Toronto scientists have discovered that the largest group of human proteins, which work as genome gatekeepers to control gene activity, are even more diverse in their roles than previously thought. The finding expands our understanding of how proteins "read" the DNA and could lead to a more accurate interpretation of individual genomes.
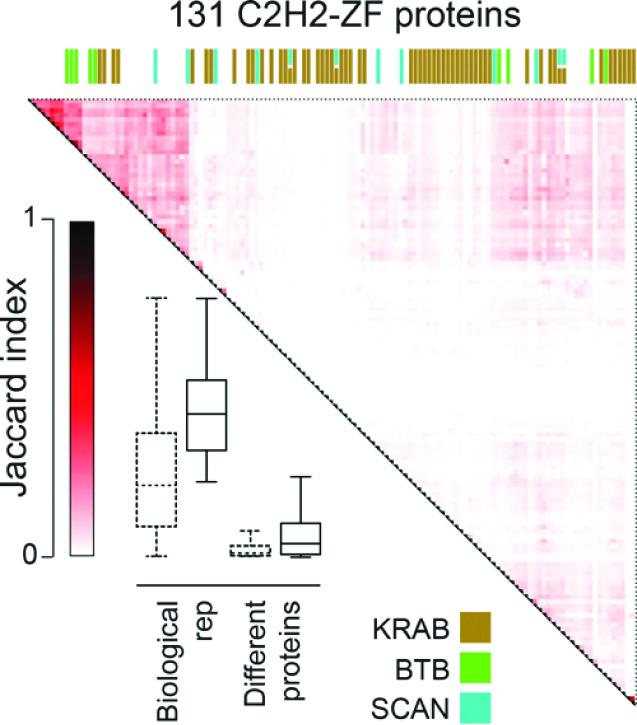
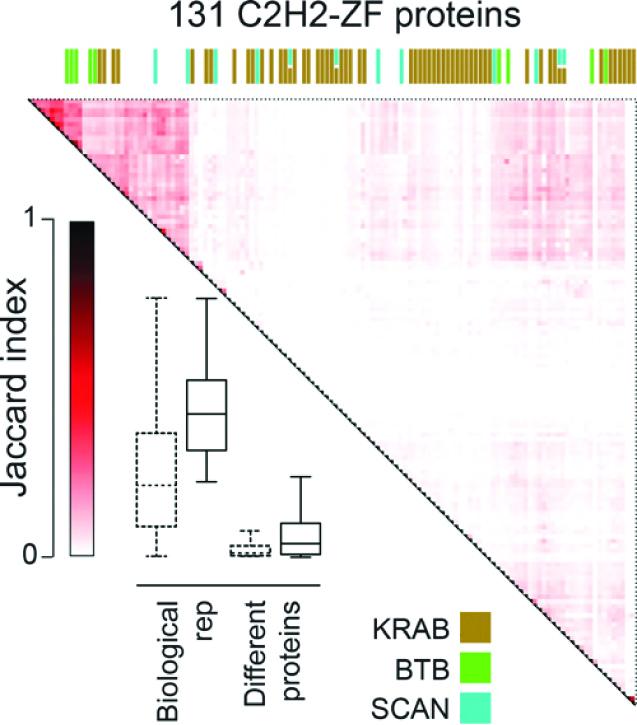
Teams led by Professors Timothy Hughes and Jack Greenblatt, of the University of Toronto's Donnelly Centre, have shown that proteins called C2H2-zinc fingers (C2H2-ZF) can control gene activity in new and surprizing ways. Reporting in the December issue of Genome Research, the researchers also reveal DNA binding sites for more than a hundred C2H2-ZFs as part of an ongoing effort to decode genome sequences that do not code for genes.
Despite being the largest group of human proteins — counting 700 members — the C2H2-ZFs are poorly understood partly because their sheer abundance and diversity make them hard to study. Yet knowing how they work is important because they help orchestrate gene activity. Of 20,000 human genes, only a subset is active in the cell at any given time. This subset determines if the cell will, say, build blood, or the brain or go haywire to become cancer.
The C2H2-ZF proteins work by directly binding the DNA to control the genes nearby. Named after their finger-like structures that, aided by zinc ions, clasp the DNA, C2H2-ZFs have previously been thought to act by stifling a wide range of genes. In a previous study that included about 40 C2H2-ZFs, the team showed that each protein recognized a unique DNA snippet as its landing site in the genome, raising the possibility that the rest of the group could be just as diverse.
This was indeed confirmed in the present study in which the teams mapped DNA binding sites, most of which were unique, this time for 131 C2H2-ZF proteins. But they also uncovered a whole new way in which the C2H2-ZF proteins can be regulated to vastly expand their job repertoire in the cell.
In addition to binding the DNA, it turned out that each C2H2-ZF can partner with a motley of other proteins that could potentially tweak its ability to switch genes on and off in a unique way. The finding upended the previous thinking in which C2H2-ZF proteins were seen as limited in their ability to bind other proteins–half of them were thought to interact with a single protein that helps them gag target genes, while the rest lack the usual molecular features that help proteins contact one another.
"Our key finding is that there's almost as much diversity in the protein-protein interactions as there is in the DNA binding sequences. It tells us that the way the C2H2-ZF proteins work is almost certainly more complicated than we would have expected," said Hughes, who is also a professor in U of T's department of molecular genetics and a fellow of the Canadian Institute for Advanced Research (CIFAR).
The kinds of proteins that C2H2-ZFs interact with suggest that their roles go beyond clamping down on genes and may even act to turn genes on or help package DNA inside the cell.
The study also shines light on how the C2H2-ZF evolved to become the largest and most diverse group of proteins we have. The DNA sequences that C2H2-ZF proteins recognize look a lot like they had come from viruses, some of which plagued our mammalian ancestors as long as 100 million years ago. This kind of viral DNA has been called "selfish DNA" because it spreads by inserting itself randomly in a host's genome.
It is thought that the C2H2-ZF proteins evolved to shut down this selfish DNA, their legion expanding to keep up with new intruders. Once the viral DNA was squashed for good, the C2H2-ZF proteins were able to take on new roles in shutting down mammalian genes. And now, this new data suggest that the C2H2-ZF proteins branched out even more than previously thought to acquire wholly unexpected functions by binding to other proteins.
Knowing how C2H2-ZFs work will give scientists a better handle on predicting which genes they control and how this may relate to disease. So far, mass genome sequencing studies have fallen short from being able to tell one's risk of common diseases, such as cancer or diabetes, because we still don't know enough about the meaning of individual differences between genomes.
"Even today, 15 years after the human genome was sequenced, if you give any piece of DNA to a geneticist and ask them what this does, generally they are unable to tell you that. But the more we learn about how human proteins recognize the DNA and what they do, the better our ability will be to interpret genome sequences and say what the significance of the variants is," said Hughes.
###
@DonnellyCentre
http://www.thedonnellycentre.utoronto.ca/
Media Contact
Jovana Drinjakovic
[email protected]
416-543-7820
@UofTNews
http://www.utoronto.ca
############
Story Source: Materials provided by Scienmag