Credit: Courtesy of the Mazmanian laboratory
The mammalian gut is warm, moist, and incredibly nutrient-rich–an environment that is perfect for bacterial growth. The communities of "good bacteria" in the gut, commonly referred to as the microbiome, are vital partners for the body, helping to digest fiber, extract nutrients, and prevent various diseases. We are all familiar with the immune responses and illnesses that ensue from bad, or pathogenic, bacteria entering the body–so, if the immune system evolved to repel microbes, then how do mammals maintain harmonious relationships with the beneficial bacteria in the gut?
Now, new research from Caltech illustrates how a particular species of beneficial bacteria actually harnesses the body's immune response so that it can settle down comfortably in the gut.
The work was done in the laboratory of Sarkis Mazmanian, Luis B. and Nelly Soux Professor of Microbiology and Heritage Medical Research Institute Investigator. A paper describing the research was published online on May 3 in the journal Science.
Led by graduate student Gregory Donaldson, researchers in the Mazmanian laboratory chose to examine a microbe called Bacterioides fragilis. The particular species is found abundantly in the large intestines of many mammals, including humans, and was previously shown by the Mazmanian lab to protect mice from certain inflammatory and neurological disorders such as inflammatory bowel disease and multiple sclerosis. Interestingly, though there are multiple strains of B. fragilis, healthy people form a long-term, monogamous relationship with only a single strain.
"Studies by other labs have shown that most people carry the same strain of B. fragilis throughout their lives," says Donaldson. "We wanted to understand at a molecular level how these bacteria are able to colonize the gut in a stable, long-term way."
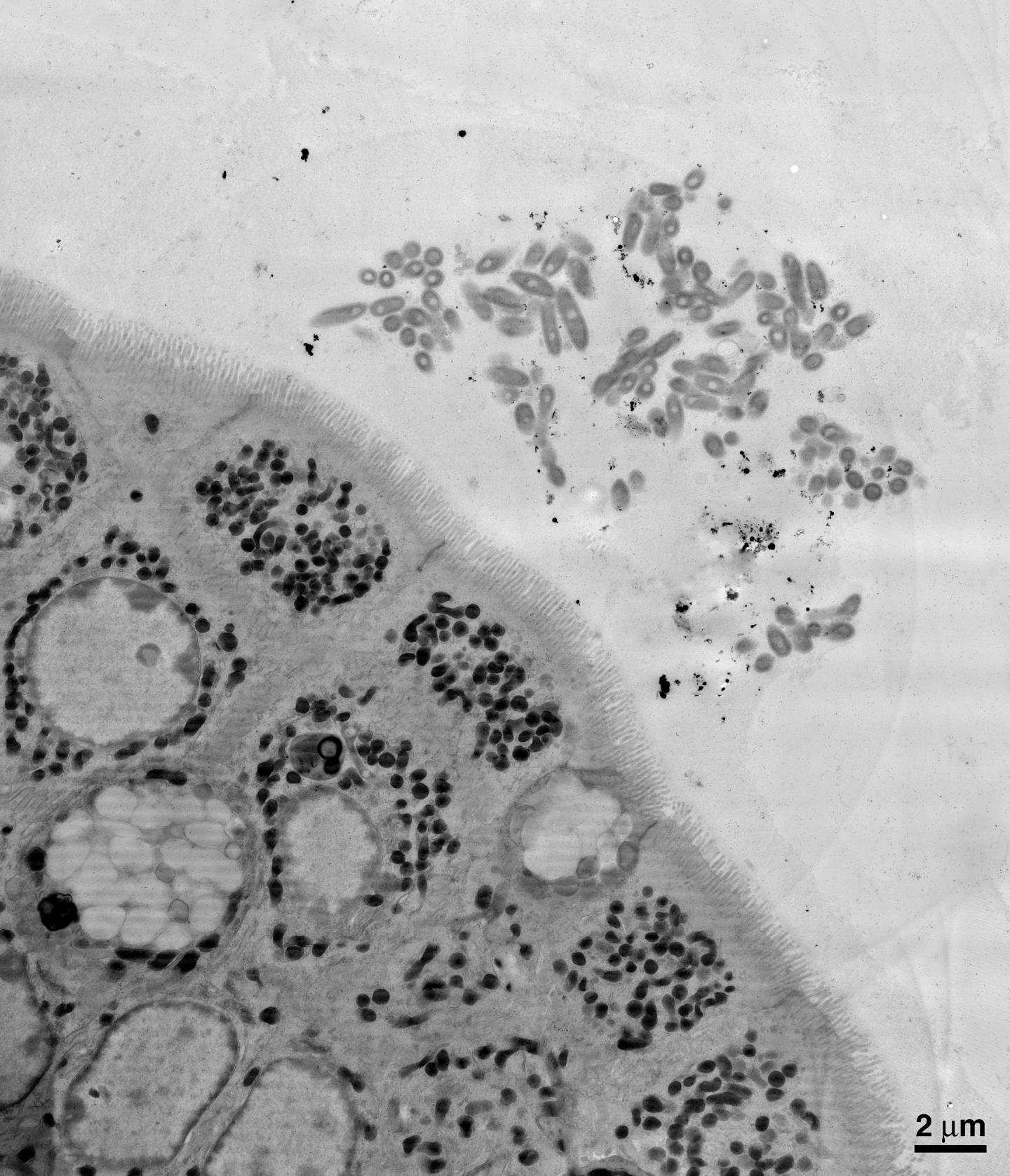
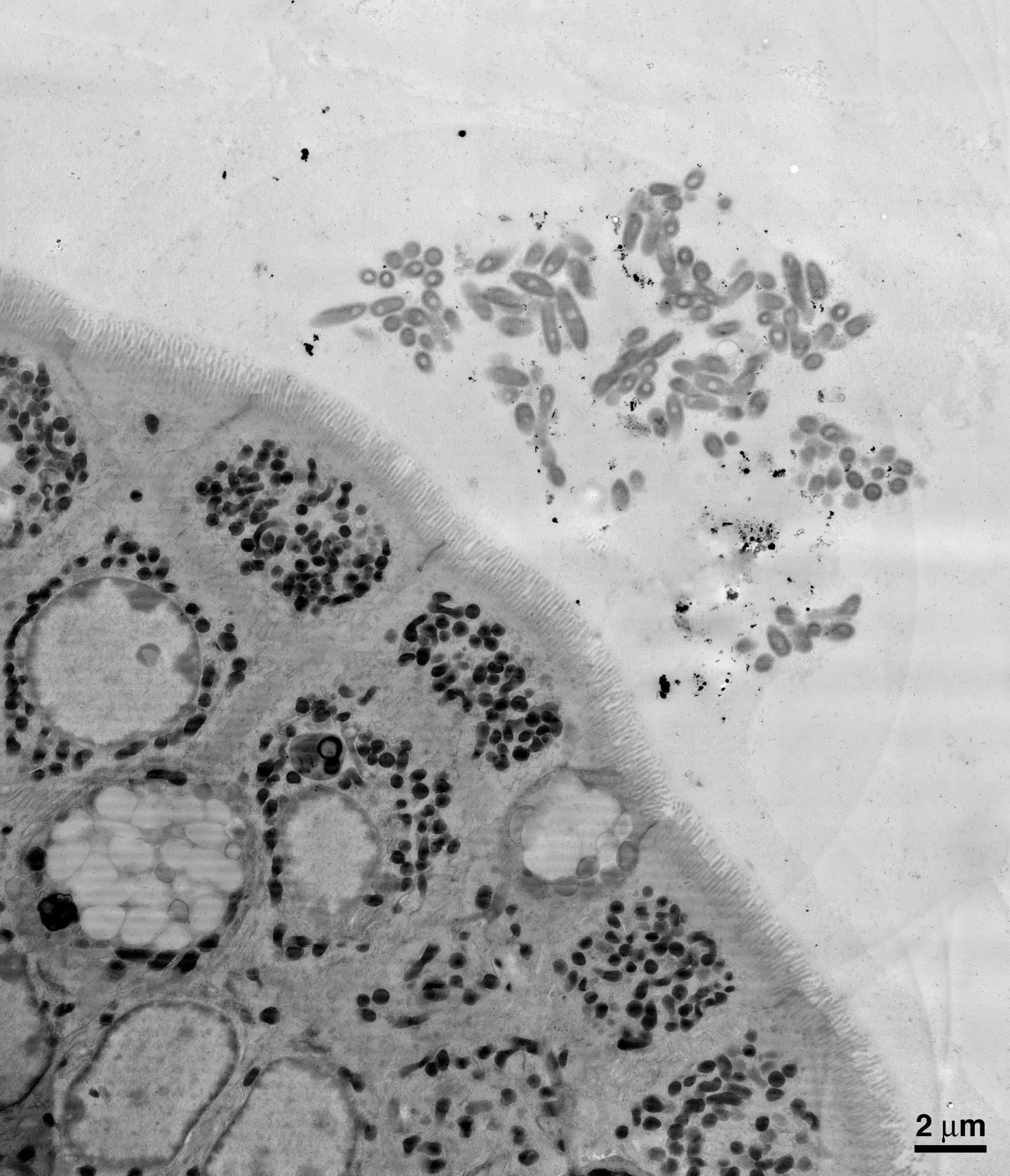
First, the researchers aimed to examine B. fragilis's symbiotic relationship with the gut by physically looking at the locations where the bacteria reside. Using electron microscopy imaging on samples of mouse intestines, the team was able to see that B. fragilis clumps together in aggregates deep within the thick layer of mucus lining the gut, nestled close to the epithelial cells that line the surface of the intestine. Donaldson and his collaborators theorized that this spatial niche is necessary for a single species to settle in and establish a stable foothold.
The team next aimed to determine what mechanisms allow B. fragilis to colonize such a niche within the gut. They found that each B. fragilis bacterium is encased in a thick capsule made of carbohydrates. The capsule is typically associated with pathogens (bad bacteria) attempting to cloak themselves from recognition by and attack from the body's immune system. Mutant bacteria lacking this capsule cannot aggregate and do not inhabit the mucosal layer. Thus, the researchers theorized that capsular carbohydrates are necessary for B. fragilis strains to monopolize their niche in the gut.
Because bacterial capsules were known to be related to an immune response in pathogenic bacteria, Donaldson and Mazmanian hypothesized that there may also be an immune response to the B. fragilis capsule. Indeed, they found that antibodies, immune proteins that grab onto and mark specific bacteria or viruses for other immune cells to engulf and destroy, were binding to the B. fragilis capsule in the intestine. One particular kind of antibody, immunoglobulin A or IgA, is found throughout the gut–in fact, it is the most abundantly produced type of antibody in humans–but its specific functions have been enigmatic.
Normally, an antibody response means imminent death to pathogenic bacteria. But curiously, IgA does not negatively affect most of the bacteria that normally live in the gut. In the case of B. fragilis, the researchers found, it actually helped the bacteria stick to epithelial cells. Furthermore, in mice that lacked IgA, the bacterium was less successful at colonizing the surface of the intestine and maintaining long-term stability.
The team believes that this IgA response to the B. fragilis capsule helps anchor the bacteria to the epithelial surface, thus providing an advantage.
"It is surprising to find that an immune response actually helps beneficial bacteria to thrive, which in turn helps the host thrive," says Donaldson. "The study of immunology has mainly been in the context of pathogenic bacteria. But there are trillions of bacteria in the gut, and most of the time none of them are making you sick. Our study shows that there is active immune recognition of these bacteria, but it helps rather than hinders them. This suggests that the immune system is more than just a defense system and antibodies are more than just weapons."
In future work, the researchers plan to study how the gut's antibody response arises in the first place and why it helps B. fragilis while other antibodies hurt bacteria. Ultimately, this work could be used to improve colonization by other beneficial bacteria, as through the use of probiotics.
"Over the past decade, many studies have profiled the gut microbiome in a variety of diseases, lifestyles, geographies, and following birth," says Mazmanian. "We've learned that the community composition of the microbiome correlates with particular conditions–for example, altered microbiome configurations may contribute to inflammatory bowel disease, autism, and Parkinson's disease. What has remained largely unknown is how a microbiome is established and maintained in the first place. Our study reveals a molecular mechanism by which specific beneficial bacteria actively promote long-term intestinal colonization by engaging and co-opting the immune system, rather than trying to evade it as pathogens do. This discovery may lead to new ways to correct microbiome imbalances, and perhaps to prevent and treat a variety of human disorders."
###
The paper is titled "Gut microbiota utilize immunoglobulin A for mucosal colonization." In addition to Donaldson and Mazmanian, other Caltech co-authors include Mark Ladinsky, electron microscopy scientist; Kristie Yu (BS '17), former undergraduate student; Bryan Yoo, graduate student; and Pamela Björkman, Centennial Professor of Biology. Additional co-authors are Jon Sanders and Rob Knight of UC San Diego, Wen-Chi Chou and Ashlee Earl of the Eli & Edythe Broad Institute of MIT and Harvard, and Margaret Conner of Baylor College of Medicine. Funding was provided by the National Institutes of Health, the National Science Foundation, and the Heritage Medical Research Institute.
Media Contact
Lori Dajose
[email protected]
626-658-0109
@caltech
http://www.caltech.edu
Original Source
http://www.caltech.edu/news/gut-bacteriums-guide-building-microbiome-82131 http://dx.doi.org/10.1126/science.aaq0926