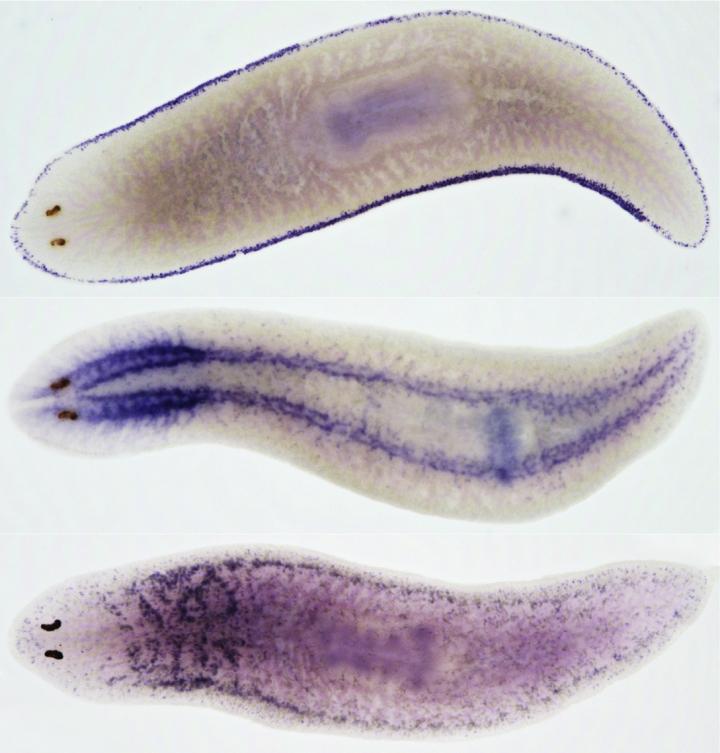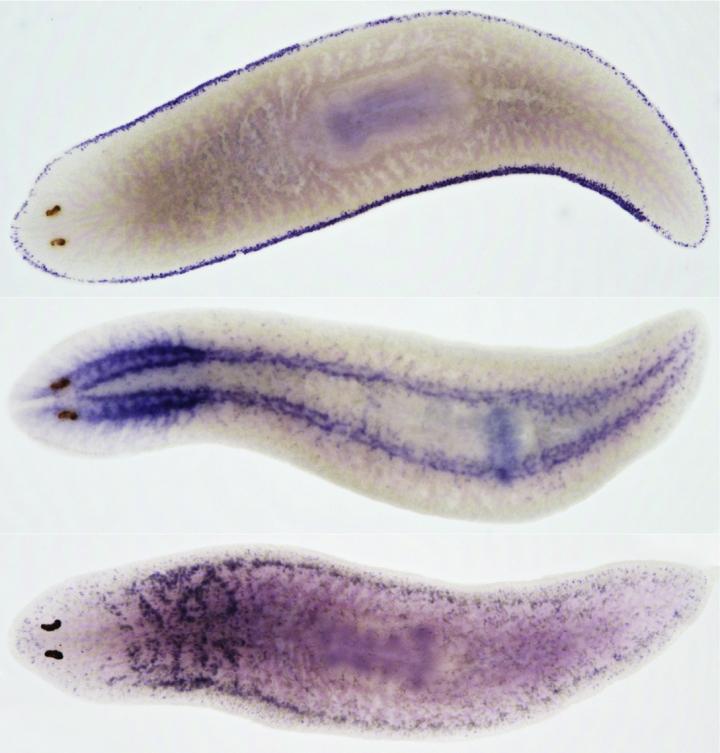
Credit: Credit: Salah Ayoub, BIMSB/MDC and Jordi Solana, BIMSB/MDC, now at Oxford Brookes University
From one stem cell to many differentiated body cells: Scientists from the MDC in Berlin, along with collaborating researchers in Munich, have published a comprehensive lineage tree of a whole adult animal in the journal Science. This was made possible by a combination of RNA and computational technologies.
How do uniform stem cells become complex body cells that serve a wide variety of functions? The differentiation of stem cells into the multiplicity of cell types in the human body is an enigma of modern medical science. An interdisciplinary group of scientists from the Max Delbrück Center for Molecular Medicine (MDC) in Berlin and the Helmholtz Center in Munich has made important progress with a comprehensive study published in the journal Science. By combining sophisticated single-cell RNA technology with nucleic acid sequencing and computational methods, the scientists were able to establish a detailed cell atlas of an entire complex adult animal, the Schmidtea mediterranea flatworm, and reconstruct the lineage tree of all identified cells. Their work also provides new insights into cellular regeneration processes. "It opens the door to powerful new approaches to study the cell composition of organs, tissues, and developmental stages and to understand the molecular mechanisms of regeneration," says Professor Nikolaus Rajewsky, head of Berlin Institute for Medical Systems Biology (BIMSB), at the MDC, and senior author of the study.
Researchers reconstructed a comprehensive lineage tree, including 23 cellular differentiation pathways
Planarians such as Schmidtea mediterranea are popular study objects because they are immortal and can completely regrow if cut into pieces. They possess a large number of adult pluripotent stem cells that constantly renew all tissues and cell types. "For understanding how cells differentiate, we need to compare gene expression profiles from cells at various differentiation stages, like the ones we find in planarians," says Dr. Mireya Plass from the MDC. "We dissociated whole animals and characterized thousands of individual cells by sequencing their messenger RNA transcripts." The scientists identified 37 different cell types, 23 of them terminally differentiated, as well as numerous stem cells and so-called progenitor cells at several levels of differentiation.
"We then applied a new computational algorithm, PAGA, developed by Dr. Alexander Wolf, and Professor Fabian Theis in Munich, to predict a lineage tree that includes all identified cell types rooted to a single stem cell group," says Dr. Christine Kocks from the MDC. This prediction was then combined with additional computational and experimental methods to build a robust tree supported by several independent lines of evidence: sequencing of purified stem cells and cells from stem cell-depleted animals; gene expression changes; unsupervised lineage tree prediction from PAGA; and the results from a recently published method, velocyto, that predicts future cell states from mRNA metabolism estimated from precursor and mature transcripts. In addition, the researchers were able to identify gene programs involved in cell differentiation processes along this tree.
The scientists also discovered several new cell types located in the parenchyma of the animals that had been previously overlooked in molecular studies and that play an important role in the regeneration of flatworms. The proportion of these cells is significantly diminished in regenerating animals, suggesting that these cells "fuel" the body remodeling process in planarians.
Additional online and interactive resources are available to the scientific community
Scientists worldwide have immediate access to the new data on flatworm biology as well as the methods used by the team of researchers. "Our work makes it possible to study stems cells and their lineages in multiple animals. It is not only an important resource for the planarian community, but also provides a cookbook for other scientists that want to perform similar studies in other organisms", says Dr. Jordi Solana, formerly MDC, now Oxford Brookes University. Via an interactive app scientists can interrogate the data and look at the changes in gene expression of specific genes during cell differentiation and regeneration. The detailed online tutorial for lineage reconstruction using PAGA and velocyto will provide a guide for future studies. "Single-cell approaches will become an indispensable method for studying developmental and regeneration biology," concludes Nikolaus Rajewsky.
###
Reference
Mireya Plass et al. (2018): "Cell type atlas and lineage tree of a whole complex animal by single-cell transcriptomics,", Science. Advance Online Publication 19.04.2018, doi:10.1126/scienc.aaq1723
About the Max Delbrück Center
The Max Delbrück Center for Molecular Medicine in the Helmholtz Association was founded in January 1992 on the recommendation of the German Council of Science and Humanities ("Wissenschaftsrat") with the goal of linking basic science to clinical research. The MDC integrated parts of three former Central Institutes of the GDR Academy of Sciences and was named for Max Delbrück, a physicist, biologist, and Nobel Prize winner. Currently the institute employs more than 1600 people from nearly 60 countries; over 1300 of those are directly involved in research. The MDC's annual budget is over 80 million Euros, along with substantial third-party funding obtained by individual scientific groups. As is the case with all Helmholtz institutes, the MDC receives 90 percent of its funding from the federal government and 10 percent from Berlin, the state where it resides. http://www.mdc-berlin.de
Media Contact
Annette Tuffs
[email protected]
49-309-406-2140
http://www.mdc-berlin.de
Related Journal Article
http://dx.doi.org/10.1126/scienc.aaq1723