Credit: Kota et. al./The Scripps Research Institute
JUPITER, FL – May 14, 2018 – Cancer is a disease often driven by mutations in genes. As researchers learn more about these genes, and the proteins they code for, they are seeking smarter drugs to target them. The ultimate goal is to find ways to stop cancer cells from multiplying out of control, thereby blocking the growth and spread of tumors.
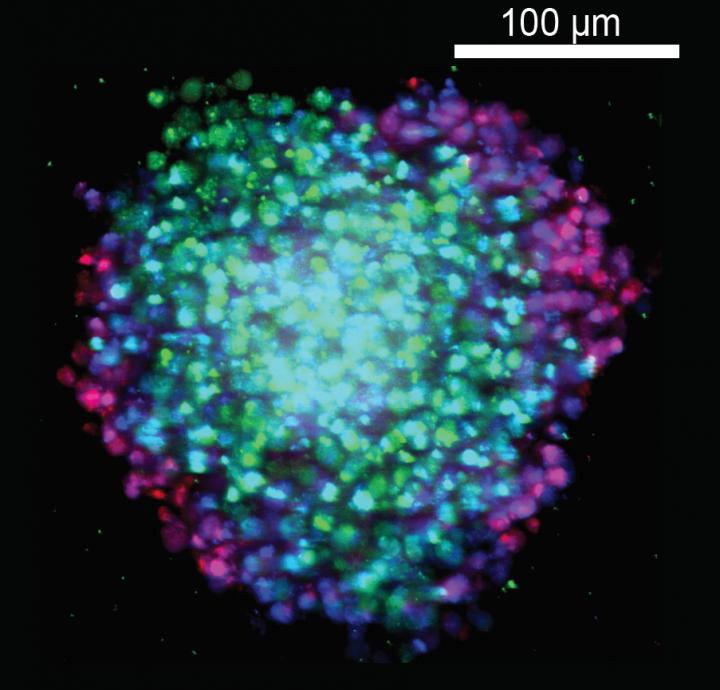
Now researchers from The Scripps Research Institute are reporting an innovative new method to screen for potential cancer drugs. The technique makes use of tiny, three-dimensional ball-like aggregates of cells called spheroids. These structures can be used to interrogate hundreds or even thousands of compounds rapidly using a technique called high throughput screening. In fact, by using this approach, the team has already identified one potential drug for an important cancer gene. The results were reported in the journal Oncogene.
"What's important about this research is that we're able to do studies using a form of cancer cells that is more physiologically relevant and better recapitulates how these cells appear in the body," says Timothy Spicer, director of Lead Identification Discovery Biology and High Throughput Screening on Scripps Research's Florida campus and one of the study's corresponding authors.
"Until now, most of the research to screen for cancer drugs has used cells that are growing flat on a plate," adds Louis Scampavia, director of HTS Chemistry and Technologies at Scripps Research and one of the study's co-authors. "With these 3-D spheroids, we emulate much more closely what's found in living tissues."
The spheroids are 100 to 600 microns in diameter-equivalent to the thickness of a few sheets of paper. In contrast to single layers of cells normally used to screen for drugs, which tend to all grow at the same rate because they get the same exposure to oxygen and nutrients, the spheroids mimic what might happen in a tumor: Some cells are on the outside and some are on the inside.
In the new paper, the researchers focused on a cancer-driving protein called KRAS. The KRAS gene and other members of the related RAS gene family are found to be mutated in nearly one-third of all cancers. They are common in lung cancer, colorectal cancer, and especially pancreatic cancer. In fact, up to 90 percent of pancreatic cancers are driven by KRAS mutations, and the investigators used pancreatic cancer cell lines for the current study.
"In the past, KRAS has been a very tricky protein to target. People have spent several decades trying, but so far there has been little success," says Joseph Kissil, PhD, professor at Scripps Research Medicine and the other co-corresponding author. "The KRAS protein is relatively small, and that's made it hard to attack it directly. But the method of screening that we used in this study allowed us to come at the question in a different way."
The investigators performed what is called a phenotypic screen, which means they were looking for drugs that had an effect on cell growth, but didn't have a preconceived idea about how they might work. "We came at this in an unbiased way," Kissil explains. "We were not trying to design something to attack a specific part of the KRAS protein. We were just looking for something that acted on some part of the pathway that's driving cell growth."
The investigators report in the new paper that they have already identified one compound that was previously not know to affect KRAS, called Proscillaridin A. The compound is similar to a class of drugs used to treat some heart conditions. Although the team says this particular drug is unlikely to be developed as a cancer treatment, it validates the approach of conducting drug screenings using spheroids. "It's unlikely we would have discovered this connection using standard 2-D methods," Scampavia says.
"From our perspective, this is a proof-of-principle study," Kissel adds. "It shows you can look at libraries of drugs that have already been approved for other diseases, and find drugs that may also work for cancer. In theory, you could use this screening method for any line of cancer cells, and any mutation you want."
"We would love to use this research to create a pipeline for new oncology drugs," Spicer concludes. "Many of the most promising compounds may be overlooked with 2-D screening. This study provides direct evidence that screening for drugs using 3-D structures of cancer cells may be more appropriate."
###
Other authors of the study, "A Novel 3-dimensional High Throughput Screening Approach Identifies Inducers of a Mutant KRAS Selective Lethal Phenotype," were Smitha Kota, Shurong Hou, William Guerrant, Franck Madoux, Scott Troutman, Virneliz Fernandez-Vega, Nina Alekseeva and Neeharika Madala of Scripps Research.
This work was supported in part by the National Cancer Institute of the National Institutes of Health (grants R33CA206949 and CA124495).
About The Scripps Research Institute
The Scripps Research Institute (TSRI) is one of the world's largest independent, not-for-profit organizations focusing on research in the biomedical sciences. TSRI is internationally recognized for its contributions to science and health, including its role in laying the foundation for new treatments for cancer, rheumatoid arthritis, hemophilia, and other diseases. An institution that evolved from the Scripps Metabolic Clinic founded by philanthropist Ellen Browning Scripps in 1924, the institute now employs more than 2,500 people on its campuses in La Jolla, CA, and Jupiter, FL, where its renowned scientists-including two Nobel laureates and 18 members of the National Academies of Science, Engineering or Medicine-work toward their next discoveries. The institute's graduate program, which awards doctoral degrees in biology and chemistry, ranks among the top ten of its kind in the nation. In October 2016, TSRI announced a strategic affiliation with the California Institute for Biomedical Research (Calibr), representing a renewed commitment to the discovery and development of new medicines to address unmet medical needs. For more information, see http://www.scripps.edu/.
Media Contact
Stacey Singer DeLoye
[email protected]
561-228-2551
@scrippsresearch
http://www.scripps.edu
Related Journal Article
http://dx.doi.org/10.1038/s41388-018-0257-5