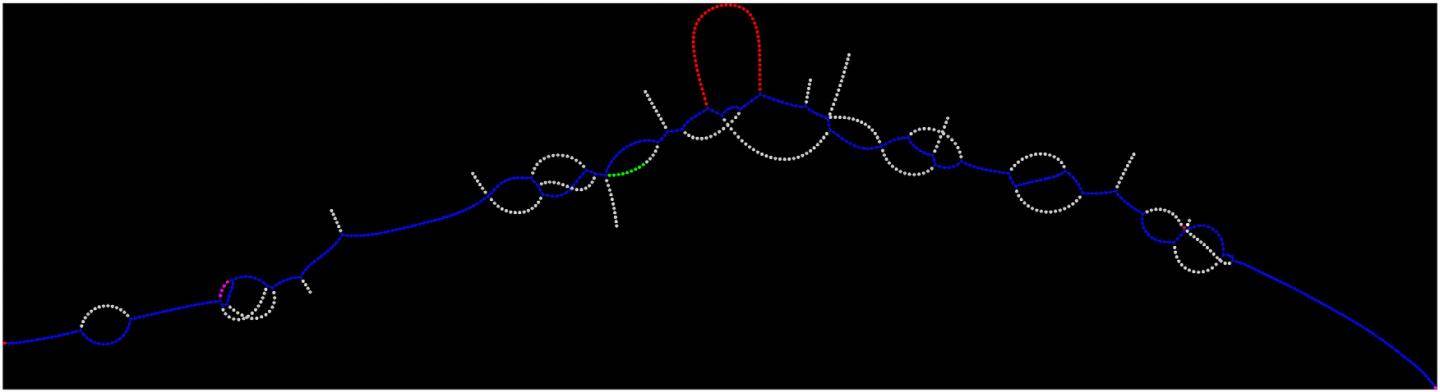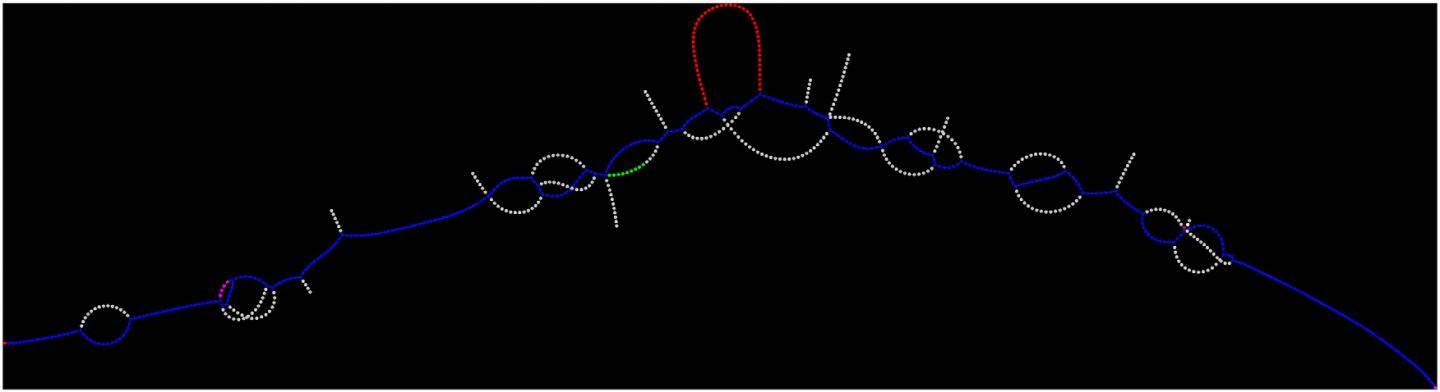
Credit: New York Genome Center
NEW YORK, NY (March 22, 2018) — A new computational method developed by researchers at the New York Genome Center (NYGC) allows scientists to identify rare gene mutations in cancer cells with greater accuracy and sensitivity than currently available approaches.
The technique, reported in today's Communications Biology from Nature Research publishing, is called Lancet and represents a major advance in the identification of tumor cell mutations, a process known as somatic variant calling.
"With its unique ability to jointly analyze the whole genome of tumor and matched normal cells, Lancet provides a useful tool for researchers to conduct more accurate genome-wide somatic variant calling," notes first author Giuseppe Narzisi, PhD, Senior Bioinformatics Scientist, NYGC.
"Reliable detection of somatic variations is of critical importance in cancer research and increasingly in the clinical setting, where identification of somatic mutations forms the basis for personalized medicine," said Michael Zody, PhD, Senior Director, Computational Biology, NYGC, and senior author of the study. "Lancet will be an important addition to the toolkit of both clinicians and researchers working to advance the field of cancer genomics and improve care for cancer patients."
To identify gene mutations in cancer cells, researchers sequence the genomes of tumor cells and normal cells. Current computational methods then involve comparing both tumor and normal to a reference genome and looking for differences unique to the tumor. Lancet instead uses an approach called micro-assembly to reconstruct the complete sequences of small regions of the genome without relying on a reference. Because the approach does not rely on a reference to identify variants, it also works well in regions of the genome where comparing reads to a reference is challenging for technical reasons. By using a data structure called a colored de Bruijn graph, Lancet jointly analyzes the tumor and normal DNA, providing greater sensitivity to find rare variants unique to the tumor while also providing greater accuracy of differentiating tumor variants from those present in healthy tissue in that individual. Using Lancet to combine the sequencing data from the normal and tumor cells represents a more powerful way of identifying mutations, Dr. Narzisi said, since users are no longer dependent on analyzing sequence data from tumor and normal cells separately.
In the study, through extensive experimental comparison on synthetic and real whole-genome sequencing datasets, the researchers demonstrated that Lancet performed better and had higher accuracy and better sensitivity to detect somatic variants compared to the most widely-used somatic variant callers.
"In our study, we show that existing tools are not that precise in scoring mutations, so that some candidate variants which were highly scored by some tools ended up being false positives," Dr. Narzisi said. "That becomes a problem when you want to prioritize which variants to validate using other technologies or you want to move forward with a clinical study. You may end up focusing on variants that do not exist."
###
To facilitate its widespread use in the scientific and medical community, Lancet is freely available for non-commercial use at https://github.com/nygenome/lancet.
About the New York Genome Center
The New York Genome Center is an independent, nonprofit academic research institution at the forefront of transforming biomedical research with the mission of advancing clinical care. A collaboration of premier academic, medical and industry leaders across the globe, the New York Genome Center has as its goal translating genomic research into the development of new treatments, therapies and therapeutics against human disease. NYGC member organizations and partners are united in this unprecedented collaboration of technology, science and medicine, designed to harness the power of innovation and discoveries to advance genomic services. Their shared objective is the acceleration of medical genomics and precision medicine to benefit patients around the world. For more information, visit our website at http://www.nygenome.org.
Member institutions include: Albert Einstein College of Medicine, American Museum of Natural History, Cold Spring Harbor Laboratory, Columbia University, Hospital for Special Surgery, The Jackson Laboratory, Memorial Sloan Kettering Cancer Center, Icahn School of Medicine at Mount Sinai, NewYork-Presbyterian Hospital, The New York Stem Cell Foundation, New York University, Northwell Health, Princeton University, The Rockefeller University, Roswell Park Cancer Institute, Stony Brook University, Weill Cornell Medicine and IBM.
Media Contact
Karen Zipern
Director of Communications
O: 646-977-7065
M: 917-415-8134
[email protected]
Media Contact
Karen Zipern
[email protected]
646-977-7065
Original Source
http://www.nygenome.org/news/novel-genomics-tool-enables-more-accurate-identification-of-rare-mutations-in-cancer-cells http://dx.doi.org/10.1038/s42003-018-0023-9