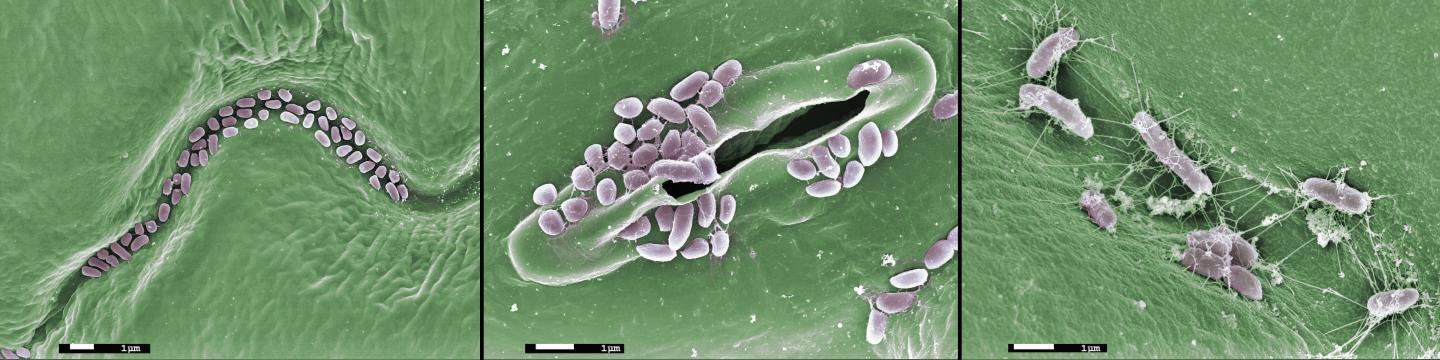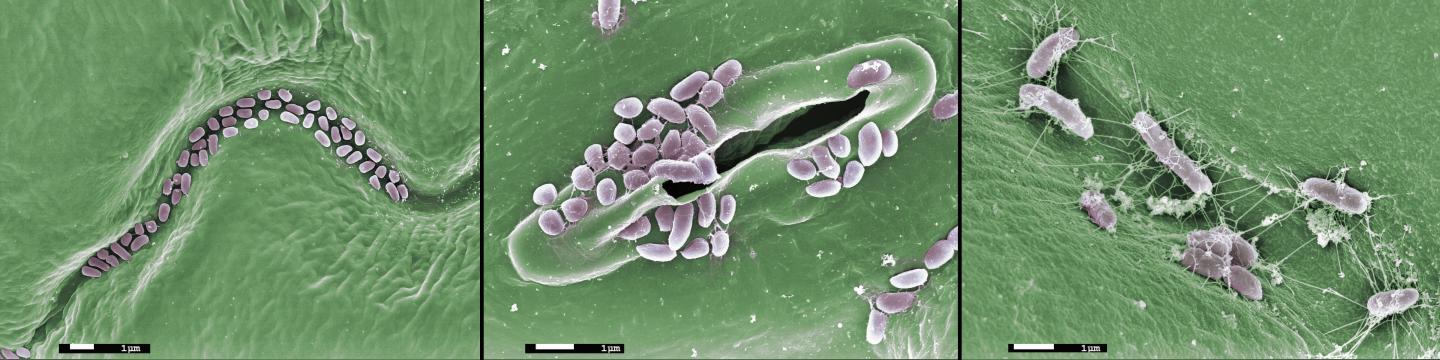
Credit: James Kremer
A coordinated effort to understand plant microbiomes could boost plant health and agricultural productivity, according to a new Perspective publishing March 28 in the open access journal PLOS Biology by Posy Busby of Oregon State University in Corvallis and colleagues at eight other research institutions.
Like humans, plants live in intimate contact with microbes, including beneficial bacteria and fungi that enhance plant growth and disease resistance. While the importance of a few individual bacterial species, such as the nitrogen-fixing rhizobia of legumes, is widely understood, relatively little is known about the structure, function, and perturbations of the complex microbial communities that surround roots and dwell on leaves.
To help understand how beneficial microbes can be harnessed in sustainable agriculture, Busby and colleagues call for a plant microbiome project modeled after the recently completed Human Microbiome Project, which provides a reference set of human microbial genome sequences to develop new tools for analyzing those genomes. The plant microbiome effort will focus on understanding relationships that impact plant growth and could aid agricultural production, including efficiency of nutrient use, stress tolerance, and disease resistance. Although some work has been done, the authors note, "there has been no coordinated effort to consolidate and translate new ideas into practical solutions for farmers."
To that end, they propose a set of five broad research priorities:
- –Develop model host microbiome systems for crop and non-crop plants. Multiple models are needed to span the range of crop plants, which include grains, vegetables, fruits, and economically important tree species. Tools for creating and working with these systems, and data derived from them, should be available in public databases.
–Define the "core microbiome," the set of organisms found in most samples of a particular set of plants, in order to identify relationships that should be prioritized for further study. Comparison of core microbiomes is likely to reveal further insights into both genetic and environmental influences on microbiome composition.
–Seek to understand the rules of microbiome assembly and resilience. As part of this goal, the authors recommend prioritizing research aimed at designing synthetic microbial communities that can successfully colonize plant organs and persist long enough in natural environments to benefit the host.
–Determine the functional mechanisms at work in agricultural microbiome interactions, to learn how these interactions promote nutrient exchange, drought tolerance, and other features of microbiome activity.
–Characterize the complex interactions among plant genotype, environmental factors, farm management strategies, and microbiome composition to learn how each influences the other. "The definition of a 'healthy' or 'beneficial' microbiome may depend on specific environmental challenges faced by the plant," the authors point out, or alternatively, there may be a "generic" microbiome whose composition promotes growth and health under a wide range of conditions.
"More so than ever before, the tools, technologies, and funding are now in place to tackle the fundamental questions in agricultural biome research," they conclude. "By encouraging the pursuit of these five key research priorities, we aim to accelerate the development of agricultural microbiome manipulations and management strategies that will increase the sustainability and productivity of global agriculture."
###
In your coverage please use this URL to provide access to the freely available article in PLOS Biology: http://dx.doi.org/10.1371/journal.pbio.2001793
Citation: Busby PE, Soman C, Wagner MR, Friesen ML, Kremer J, Bennett A, et al. (2017) Research priorities for harnessing plant microbiomes in sustainable agriculture. PLoS Biol 15(3): e2001793. doi:10.1371/journal.pbio.2001793
Funding: National Science Foundation http://www.nsf.org (grant number 1519383). Received by PEB and CS. The funder had no role in study design, data collection and analysis, decision to publish, or preparation of the manuscript. National Science Foundation http://www.nsf.org (grant number 1314095). Received by PEB. The funder had no role in study design, data collection and analysis, decision to publish, or preparation of the manuscript.
Competing Interests: The authors have declared that no competing interests exist.
Media Contact
PLOS Biology
[email protected]