Credit: The Sleiman Lab/McGIll University
McGill University researchers have chemically imprinted polymer particles with DNA strands – a technique that could lead to new materials for applications ranging from biomedicine to the promising field of "soft robotics."
In a study published in Nature Chemistry, the researchers describe a method to create asymmetrical polymer particles that bind together in a spatially defined manner, the way that atoms come together to make molecules.
Although polymers are used in everything from clothing and food packaging to 3D printing and electronics, most self-assembled polymer structures have been limited to symmetrical forms such as spherical or cylindrical shapes. Recently, however, scientists have focused on creating non-symmetrical polymer structures — for example 'Janus' particles with two different 'faces' — and they are starting to discover exciting new applications for these materials. One example: robotics made with soft, flexible structures that can change shape in response to external stimuli.
The method described in the Nature Chemistry paper "introduces a programmable level of organization that is currently difficult to attain in polymer chemistry," says McGill Chemistry professor Hanadi Sleiman, senior author of the study. "Chemically copying the information contained in DNA nanostructures offers a powerful solution to the problem of size, shape and directional control for polymeric materials."
Using DNA cages as molds
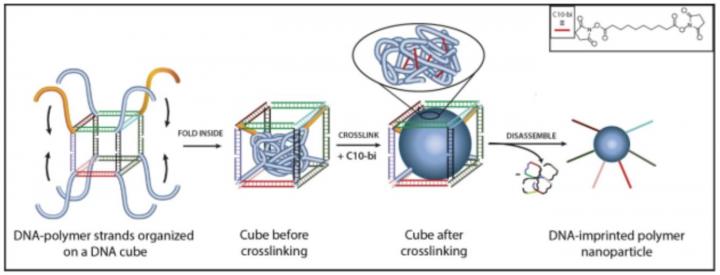
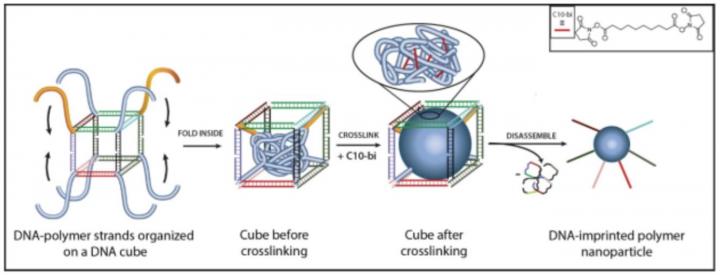
The new study builds on a technique developed in 2013 by Sleiman's research group to make nanoscale "cages" from strands of DNA, and stuff them with lipid-like polymer chains that fold together into a ball-shaped particle that can contain cargo such as drug molecules.
To take that nano-engineering feat a step further, Sleiman and her PhD student Tuan Trinh teamed up with colleagues at the University of Vermont and Texas A&M University at Qatar. Together, the researchers developed a method to imprint the polymer ball with DNA strands arranged in pre-designed orientations. The cages can then be undone, leaving behind DNA-imprinted polymer particles capable of self-assembling – much like DNA, itself – in pre-designed patterns. Because the DNA cages are used as a 'mold' to build the polymer particle, the particle size and number of molecular units in the polymer can be precisely controlled, says Sleiman, who holds the Canada Research Chair in DNA Nanoscience.
The asymmetrical polymer structures could be used eventually in a range of applications, the researchers say. One potential example: multi-compartment polymer particles, with each compartment encapsulating a different drug that could be delivered using different stimuli at different times. Another possibility: porous membranes that are asymmetric, so they direct molecules along specific paths to separate then.
###
Funding for the research was provided by the Natural Sciences and Engineering Research Council of Canada, the Canadian Institutes for Health Research, the Centre for Self-Assembled Chemical Structures, the Qatar Research Foundation, and the Canada Research Chairs Program.
"DNA-imprinted polymer nanoparticles with monodispersity and prescribed DNA-strand patterns," Tuan Trinh et al, Nature Chemistry, published online Dec. 4, 2017. doi:10.1038/nchem.2893 https://www.nature.com/articles/nchem.2893
Video: Researchers at McGill University, the University of Vermont and Texas A&M University in Qatar have developed a method to chemically imprint polymer particles with DNA strands. This approach could lead to new materials for applications ranging from biomedicine to "soft robotics." This computer simulation shows the first step of this process, in which the polymer chains associate together and fold inside the DNA cage.
CREDIT: The Li Research Group/University of Vermont
Contact:
Prof. Hanadi Sleiman
McGill University, Department of Chemistry
[email protected]
Chris Chipello
Media Relations Office, McGill University
514-398-4201
[email protected]
http://www.mcgill.ca/newsroom/
Tweets by mcgillu
Media Contact
Chris Chipello
[email protected]
514-398-4201
@McGillU
http://www.mcgill.ca
Related Journal Article
http://dx.doi.org/10.1038/nchem.1745