Credit: PeerJ
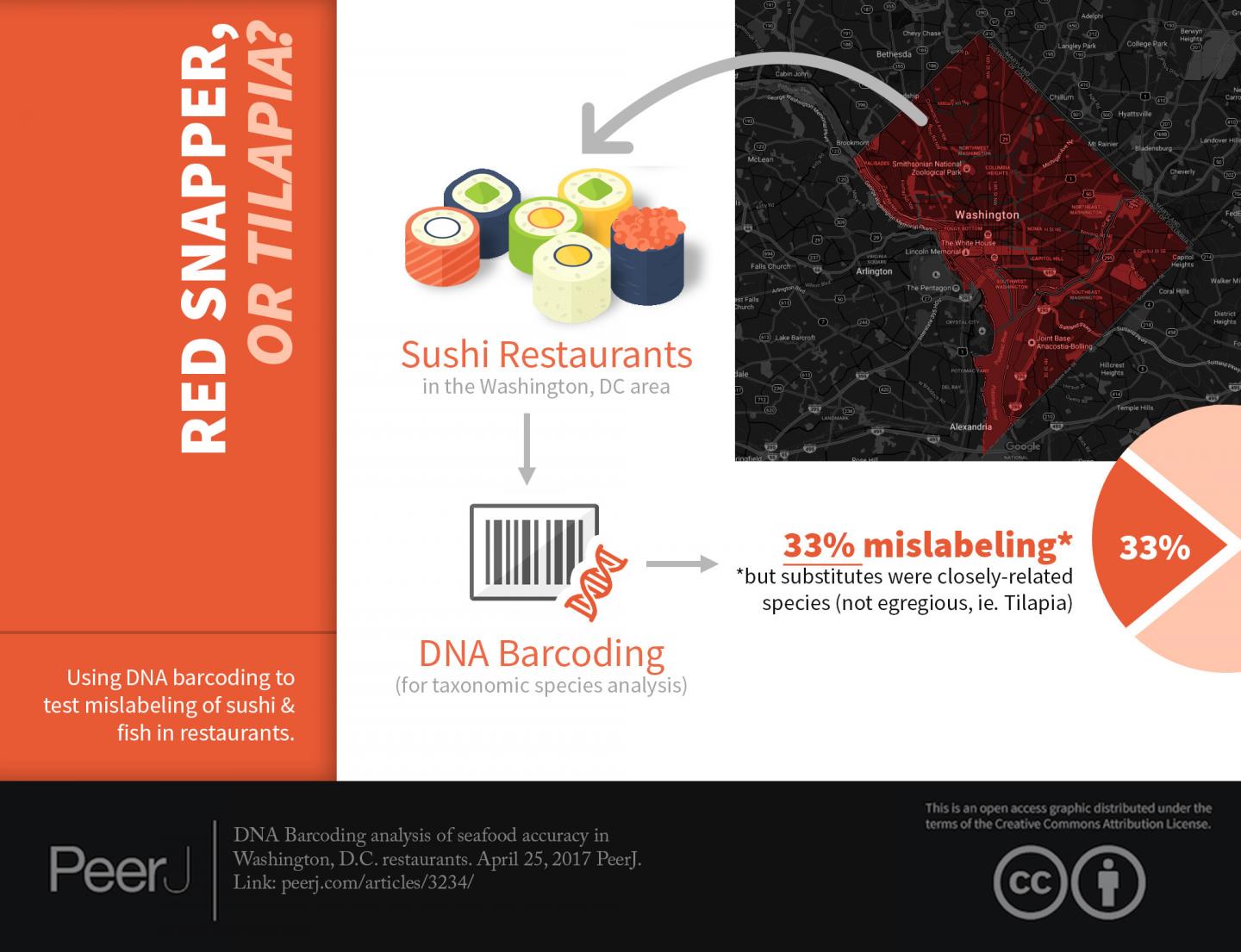
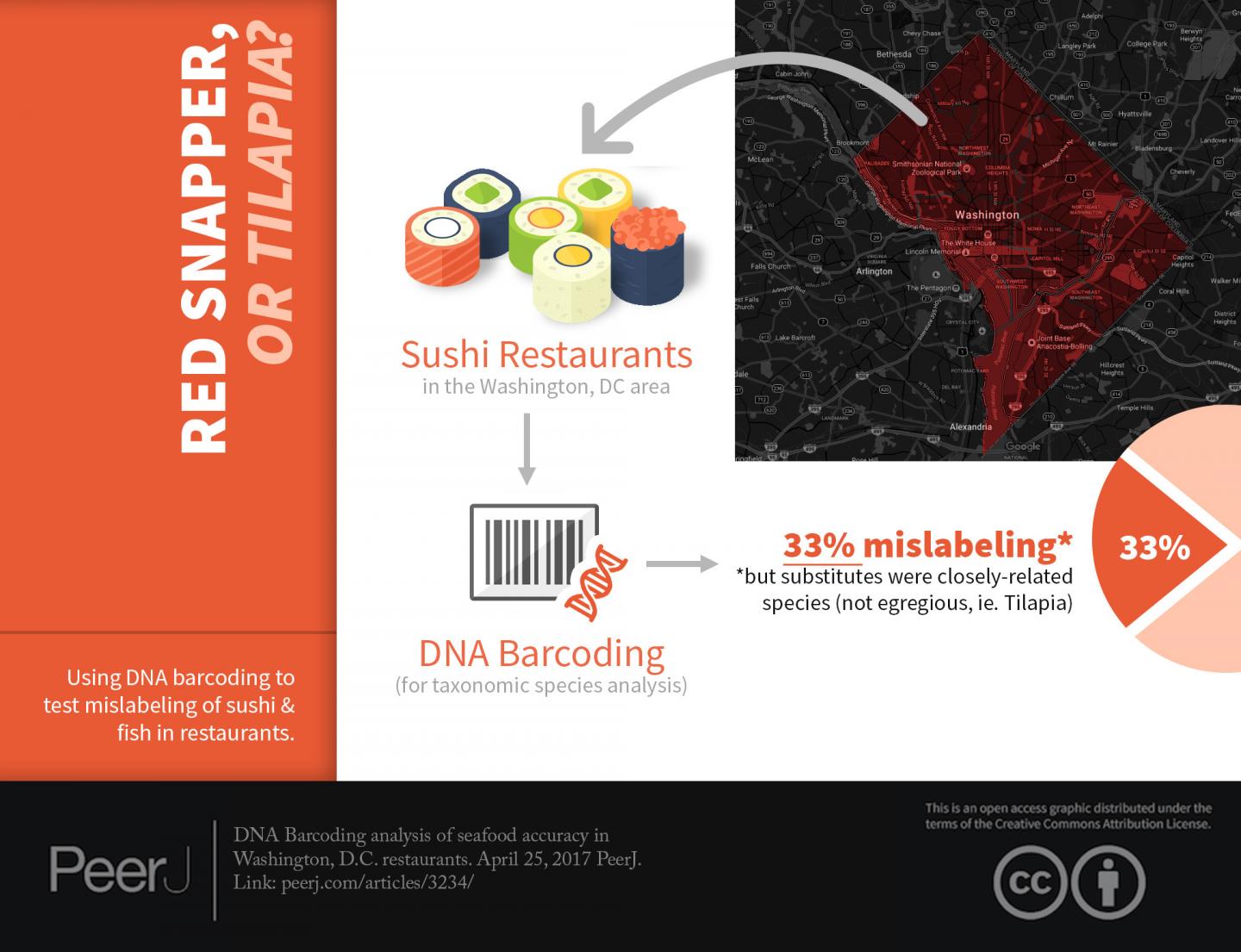
WASHINGTON, DC (April 25, 2017)– Scientists at the George Washington University used a powerful genetic technique to test seafood dinners sold in six District restaurants and found 33 percent had been mislabeled–although in most cases with species that are either closely related or considered acceptable alternatives for menu listing.
Previous studies in other cities have shown widespread seafood substitution in which consumers are sold a completely different fish or sushi from the one listed. Those studies have indicated that seafood may be mislabeled as often as 26 to 87 percent of the time.
And in egregious cases, an unsuspecting diner is sold an expensive Tuna that is actually a completely different species of fish, often one that is much cheaper or on the endangered species list, says Keith Crandall, PhD, director of the Computational Biology Institute at George Washington University's Milken Institute School of Public Health (Milken Institute SPH) and leader of the new study , which was published today in PeerJ.
Crandall and his team wondered if DC diners were getting what was listed on the menu. To find out, his team used DNA barcoding to test 12 seafood samples purchased in six restaurants and found that all in all they were doing a pretty good job.
"Diners that ordered tuna got tuna–although maybe a slightly different type of tuna," says Crandall. "We didn't see the kind of outright seafood fraud that has been reported in other cities."
For this study, Crandall recruited GW graduate student David Stern to embark on a dream assignment. Stern and his wife visited six popular DC eateries and ordered seafood dinners that caught their fancy. Not only did the two get to dine out at restaurants offering up such tasty fare like Crusted Albacore Tuna or Pesto Chilean Sea bass but Crandall instructed them to bring back the bills–he'd pick up the tab.
Stern and his wife ordered 12 seafood items on the menu but before diving into their food, they snipped off a small sample, stored it in a test tube, and brought it back to the lab. Stern then used DNA barcoding to identify a telltale region of the Cytochrome Oxidase I mitochondrial gene. This technique compares the seafood sample with a database of DNA barcodes from known species to identify it.
Of the 12 samples the research team found four menu items, one "Chilean Sea Bass" two "Tuna" and one "Rock Shrimp" had been mislabeled.
Other studies have found many instances in which restaurants swap out the item listed on the menu for a species that is endangered or threatened. But the GW team found only one sample with a conservation concern. The team's DNA barcoding analysis of the "Everything Tuna" sample identified it as Thunnus obesus, a species of tuna that is listed as "Vulnerable" by the International Union for Conservation of Nature's Redlist.
The substitutions in this study were all closely related species to the menu item or a legally acceptable swap, Crandall notes. Still, swapping out seafood does a disservice to customers who are either trying to avoid species that are endangered or are paying a higher price for a species they think is a delicacy, he says.
For example, the researchers found that one DC restaurant had listed Rock Shrimp on the menu but DNA barcoding showed it was actually serving Whiteleg shrimp. Whiteleg shrimp are typically found in aquaculture farms and are not as flavorful as deep-water Rock Shrimp.
This study, and others like it, cannot pinpoint where the swap occurs. Crandall says that buyers for restaurants can inadvertently purchase mislabeled seafood. For example, substitution of a cheaper fish for a more expensive species can happen anywhere along the food chain from the fishing boat to the fishmonger or the restaurant, he says.
In response to past reports of seafood fraud, DC and other cities passed legislation aimed at protecting the consumer. In the District, the Consumer Protection Act allows patrons to purchase samples of food or other goods for the express purpose of testing their identity.
"For the most part, our study found that DC diners with a craving for seafood are getting what they paid for." Crandall says.
###
About Milken Institute School of Public Health at the George Washington University: Established in July 1997 as the School of Public Health and Health Services, Milken Institute School of Public Health is the only school of public health in the nation's capital. Today, more than 1,900 students from 54 U.S. states and territories and more than 50 countries pursue undergraduate, graduate and doctoral-level degrees in public health. The school also offers an online Master of Public Health, MPH@GW, and an online Executive Master of Health Administration, MHA@GW, which allow students to pursue their degree from anywhere in the world.
Media Contact
Kathy Fackelmann
[email protected]
202-994-8354
@GWpublichealth
http://publichealth.gwu.edu/
############
Story Source: Materials provided by Scienmag