In a groundbreaking study published in Molecular Diversity, researchers have made significant strides in the battle against malaria by unveiling new inhibitors targeting the Plasmodium falciparum dihydrofolate reductase-thymidylate synthase (PfDHFR-TS). This dual enzyme is critical for the survival and proliferation of the malaria-causing parasite, and its inhibition is a strategic approach in malaria treatment. By tapping into the rich reservoir of natural compounds provided through the ConMedNP library, the research team, led by Haiwang Djefoulna, has adopted a multi-computational approach to identify promising candidates for drug development.
The escalating rates of malaria resistance to conventional treatments have necessitated urgent innovation in the pharmaceutical landscape. Researchers have long understood that the structural uniqueness of Plasmodium falciparum presents a formidable challenge, often thwarting the effectiveness of existing therapies. This study targeted PfDHFR-TS, which is pivotal in the parasite’s metabolic pathway, ultimately interfering with folate synthesis. By carefully selecting compounds that show potential to inhibit this enzyme, the team opens the door to new avenues in antimalarial drug design.
Employing a sophisticated computational modeling strategy, the researchers utilized virtual screening methods to sift through an extensive array of natural compounds available in ConMedNP. This innovative method allows scientists to predict the interactions between the inhibitors and the target enzymes with a high degree of accuracy. The multi-computational approach not only accelerates the screening process but also enhances the precision of identifying potential inhibitors, a critical component given the vast chemical diversity in natural products.
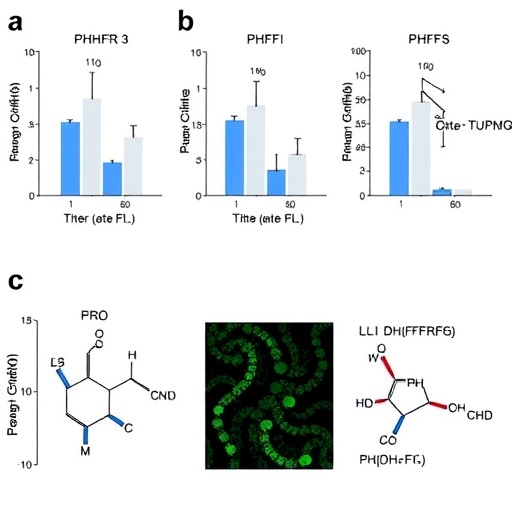
The research team performed extensive docking simulations to evaluate how well each candidate compound could bind to the active site of PfDHFR-TS. These simulations are crucial in gauging the efficacy of the compounds, as the strength and nature of binding can determine the potential success of a therapeutic agent. By analyzing the binding affinities, the researchers were able to rank the compounds and narrow down their options to the most promising candidates for further investigation.
In addition to docking studies, the researchers incorporated molecular dynamics simulations to further validate the stability and viability of the binding interactions over time. These simulations provide invaluable insights into how the compounds behave in conditions that mimic physiological environments, offering a glimpse into their potential real-world performance. This level of analysis is essential in assessing whether a compound can not only bind effectively but also endure the dynamic conditions present within a biological system.
The results of the study revealed several natural compounds that exhibited notable inhibitory activity against PfDHFR-TS. Among these, the most promising candidates were those that demonstrated strong binding affinities, illustrating their potential as viable therapeutic options. The identification of these candidates is a stepping stone towards the chemical optimization phase, where medicinal chemistry techniques can further enhance their properties and efficacy.
Beyond just identifying new inhibitors, this research underscores the importance of exploring natural compounds as a source of new pharmacological agents. The intricate chemistry and varied structural features of natural products often provide unique mechanisms of action that synthetic compounds might lack. By leveraging the biodiversity of natural compounds, researchers can potentially uncover novel solutions to chronic infectious diseases like malaria that continue to threaten global health.
The findings from this study have broad implications for future malaria treatment strategies. As resistance patterns evolve, the introduction of novel inhibitors targeting the PfDHFR-TS enzyme could play a significant role in revitalizing treatment protocols. Additionally, the research methodology exemplifies a shifting paradigm in drug discovery, where computational approaches are increasingly integral to the screening process.
As the scientific community continues to grapple with the dual challenge of malaria and drug resistance, studies like this are a beacon of hope. They not only contribute to the understanding of malaria biochemistry but also pave the way for the development of more effective and sustainable treatment options. The integration of computational techniques in drug discovery heralds a new era for researchers, enabling them to navigate complex biochemical landscapes and enhance the translational potential of their discoveries.
In conclusion, the work conducted by Djefoulna and colleagues represents a significant leap forward in the quest for effective malaria treatments. Their innovative approach, grounded in multi-computational methodologies, exemplifies how technology can reshape traditional drug discovery paradigms. As these findings move forward, they hold the potential to not only combat malaria more effectively but also inspire further exploration into the vast world of natural compounds for therapeutic applications.
The road ahead is one marked by continuous exploration, refinement, and innovation. As researchers continue to uncover new compounds from various sources, the hope is that effective therapeutic strategies will emerge, providing a means to control and ultimately eradicate this pervasive disease.
Subject of Research: Discovery of novel Plasmodium falciparum PfDHFR-TS inhibitors from ConMedNP natural compounds
Article Title: Discovery of novel Plasmodium falciparum PfDHFR-TS inhibitors from ConMedNP natural compounds: a multi-computational approach.
Article References:
Haiwang Djefoulna, V.H., Atiya Atiya, M., Fifen, J.J. et al. Discovery of novel Plasmodium falciparum PfDHFR-TS inhibitors from ConMedNP natural compounds: a multi-computational approach.
Mol Divers (2025). https://doi.org/10.1007/s11030-025-11356-7
Image Credits: AI Generated
DOI: 10.1007/s11030-025-11356-7
Keywords: Plasmodium falciparum, PfDHFR-TS inhibitors, natural compounds, computational modeling, drug discovery, malaria, resistance, multi-computational approach, docking simulations, molecular dynamics, therapeutic options, medicinal chemistry.
Tags: antimalarial drug developmentcomputational modeling in drug discoveryConMedNP library utilizationenzyme inhibitors for malariafolate synthesis interruptioninnovative malaria therapiesmalaria resistance strategiesmulti-computational approach in researchnatural compounds for malariaPfDHFR-TS inhibitorsPlasmodium falciparum treatmentvirtual screening methods in pharmacology