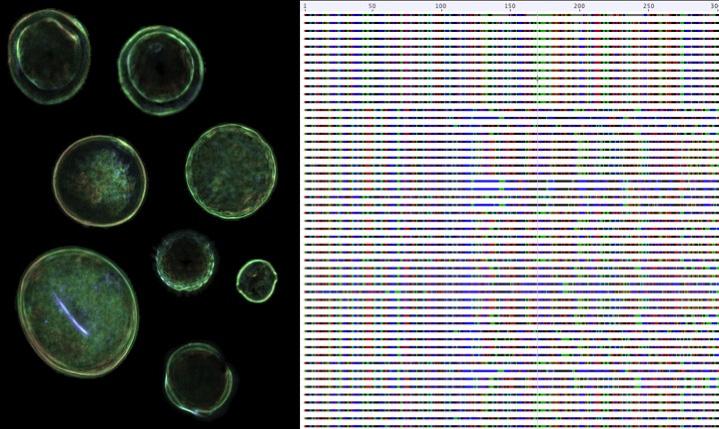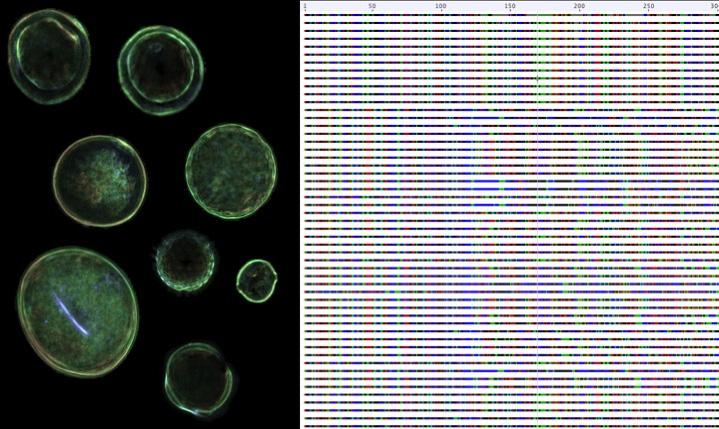
Credit: Dr. Karen L. Bell
The ability to identify individual plant species from tiny amounts of material has a surprising range of uses, from monitoring bee populations to assessing the contents of food and nutritional supplements, as well as working out what a herbivore had for breakfast. Classifying fragments of plants can be tricky, so researchers at Emory University have developed a new database of genetic information that can be used with the latest DNA sequencing technologies to improve the accuracy of plant identification.
Genetic barcodes are regions of variable DNA that can be used to identify a species by comparing its unique barcode sequence to a database of known sequences from thousands of plants. Recent advances in high-throughput DNA sequencing mean that multiple species in a mixed sample can now be distinguished and analyzed at the same time. This process, DNA metabarcoding, saves researchers the painstaking task of separating the different plant species before sequencing their DNA. Described in a new paper published in Applications in Plant Sciences , Dr. Karen Bell and colleagues from the Department of Environmental Science at Emory University used publicly available data to develop a library of sequences of the rbcL gene, a popular barcode in plants, for use in DNA metabarcoding studies.
Bell's work builds on the development of the first DNA metabarcoding database for plants, containing sequences of the ITS2 barcode from over 72,000 species. By combining ITS2 and rbcL information, the team was able to accurately identify more species from a mixed sample of pollen grains, improving the resolution and accuracy of the DNA metabarcoding technique.
The rbcL gene is a useful barcode because it codes for part of the key photosynthesis enzyme ribulose bisphosphate carboxylase (RuBisCo), so it is present in virtually all plant species. One section of its DNA sequence is very variable between species, making it ideal for DNA barcoding. Several barcoding regions have been developed in plants over the past decade, but rbcL is particularly suited to new technologies. Bell elaborates, "We chose rbcL because the length of the gene is readily applied to modern high-throughput sequencing methods." The new rbcL library contains sequences from over 38,400 plant species, around 9% of all seed plants on Earth.
The rapid innovations in high-throughput DNA sequencing have left data analysis methods behind, but the development of the rbcL and ITS2 databases means that DNA metabarcoding can be used to identify plants faster and more accurately than ever before. Using the combined rbcL and ITS2 metabarcodes, Bell and her team were able to identify eight of the nine plant species in a mixture of pollen grains – more than could be identified using the rbcL or ITS2 barcodes separately. If a species is not included in the reference library, it cannot be identified by DNA barcoding, so more sequences from the estimated 450,000 species of flowering plants must be added to make these databases more comprehensive.
Bell and her colleagues tweaked the DNA metabarcoding bioinformatics pipeline to make it capable of using additional DNA barcodes once their databases have been developed. This should further improve the barcoding accuracy because, explains Bell, "The more genetic markers available, the greater the chance of genetic identification." As the cost of genome sequencing comes down, researchers won't be restricted to scanning the barcodes of small fragments of DNA either: "At some point in the future, we'll be doing DNA barcoding using whole plant genomes. The laboratory technology is available, but currently we don't have enough complete plant genomes to make the databases."
###
This study received funding and/or support from the U.S. Army Research Office and the Emory Integrated Genomics Core (EIGC).
Karen L. Bell, Virginia M. Loeffler, and Berry J. Brosi. 2017. An rbcL reference library to aid in the identification of plant species mixtures by DNA metabarcoding. Applications in Plant Sciences 5(3): 1600110. doi:10.3732/apps.1600110
Applications in Plant Sciences (APPS) is a monthly, peer-reviewed, open access journal focusing on new tools, technologies, and protocols in all areas of the plant sciences. It is published by the Botanical Society of America, a nonprofit membership society with a mission to promote botany, the field of basic science dealing with the study and inquiry into the form, function, development, diversity, reproduction, evolution, and uses of plants and their interactions within the biosphere. APPS is available as part of BioOne's Open Access collection.
For further information, please contact the APPS staff at [email protected].
Media Contact
Beth Parada
[email protected]
@Botanical_
http://www.botany.org
############
Story Source: Materials provided by Scienmag