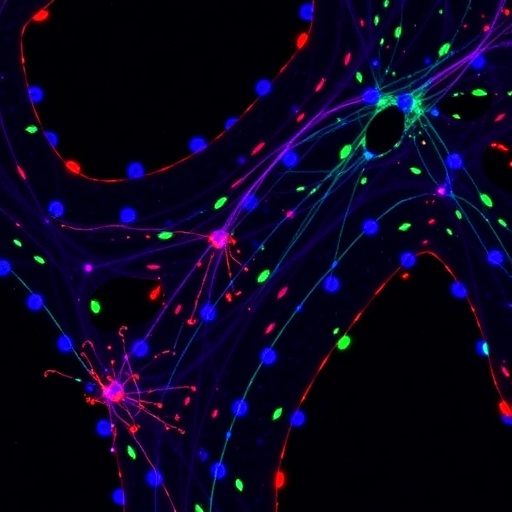
In the relentless pursuit to unravel the enigmatic mechanisms underlying Alzheimer’s disease, a groundbreaking study has illuminated the dynamic behavior of microglial cells within the brain, revealing an intricate interplay between spatial localization and epigenetic regulation. Microglia, the resident immune cells of the central nervous system, have emerged as pivotal players not only in maintaining neural homeostasis but also in mediating neuroinflammatory responses characteristic of neurodegenerative disorders. This new research, spearheaded by Ardura-Fabregat and colleagues, delves deep into the chromatin architecture that defines discrete microglial states, providing an unprecedented view into how regional heterogeneity impacts their function in a mouse model of Alzheimer’s disease.
The study presents a transformative approach by integrating spatial transcriptomics with chromatin accessibility profiling, a method that sheds light on how microglial identity and activity are sculpted by their anatomical niche within the brain. Traditionally, microglia were understood primarily through bulk molecular analyses that obscured the nuances of localized cellular states. By contrast, this research harnesses cutting-edge single-cell techniques to chart the epigenomic landscapes that underpin microglial heterogeneity, unveiling signatures that correspond with disease progression and spatial distribution.
Central to the findings is the revelation that microglia occupy distinct niches characterized by unique chromatin accessibility patterns, which in turn dictate gene expression programs tailored to local environments. These spatially defined microglial states exhibit differential responses to amyloid pathology, a hallmark of Alzheimer’s disease, suggesting that their epigenetic configuration primes them for either protective or pathological roles. Such spatial coding of microglial states offers a profound lens into the cellular choreography orchestrated in the diseased brain, challenging the conventional notion of microglia as a homogenous cell population.
.adsslot_i4hLFNyzSK{ width:728px !important; height:90px !important; }
@media (max-width:1199px) { .adsslot_i4hLFNyzSK{ width:468px !important; height:60px !important; } }
@media (max-width:767px) { .adsslot_i4hLFNyzSK{ width:320px !important; height:50px !important; } }
ADVERTISEMENT
The researchers employed a murine Alzheimer’s disease model that recapitulates key pathological features, including amyloid plaque deposition and neuroinflammation, thereby enabling the study of microglial dynamics in a controlled and relevant in vivo context. Through meticulous dissection and cell-sorting techniques, microglia were isolated from different brain regions, followed by comprehensive assays of chromatin accessibility using methods akin to ATAC-seq. This approach disclosed how epigenomic landscapes vary across anatomical boundaries and disease states, revealing region-specific enhancer and promoter usage that correlates with distinctive cellular phenotypes.
One of the seminal insights from the study is the identification of microglial subpopulations that exhibit either an activated or a homeostatic signature, demarcated not just by gene expression but by the accessibility of regulatory elements within their genome. In particular, disease-associated microglia (DAM) states displayed heightened chromatin openness at loci linked to immune function and phagocytosis, delineating a molecular framework for their reactive phenotype. Conversely, microglia in unaffected brain regions preserved chromatin landscapes supportive of surveillance and maintenance roles, highlighting the adaptability of these cells in response to local pathological cues.
Beyond mapping these discrete states, the study explores the temporal evolution of microglial chromatin landscapes as Alzheimer’s pathology progresses. Microglial populations near plaques exhibit a dynamic shift in accessibility patterns, indicating an epigenetic reprogramming that is potentially reversible or targetable by therapeutic interventions. These findings spark a compelling paradigm wherein modulating chromatin states could influence microglial function and alter disease trajectories, a prospect that brings epigenomics to the forefront of neurodegenerative research.
Another layer of complexity is added by the interplay between microglia and their microenvironment, including neurons, astrocytes, and the extracellular matrix. The researchers draw attention to the possibility that signals emanating from stressed or dying neurons may remodel microglial chromatin landscapes, inducing state transitions that either exacerbate or mitigate neurodegeneration. This bidirectional communication underscores the importance of spatial context, where proximity to pathological structures orchestrates microglial phenotypes through fine-tuned chromatin remodeling.
The application of spatially resolved epigenomics marks a significant leap forward in understanding Alzheimer’s disease, moving beyond static snapshots to capture the fluidity of microglial states in situ. This methodology captures the complexity and diversity of cellular responses embedded within the brain’s architecture, a feat previously unattainable with traditional bulk analyses. The ability to dissect regional microglial programs at the chromatin level opens new avenues for personalized and spatially informed therapeutic strategies.
Intriguingly, the study also hints at evolutionary conservation of certain microglial regulatory elements, suggesting that findings in the mouse model may translate to human pathology. The researchers posit that deciphering conserved enhancer landscapes could facilitate the discovery of universal targets for modulating microglial activity across species. This cross-species insight reinforces the relevance of spatial epigenetics in bridging preclinical models with human clinical contexts.
Moreover, the research emphasizes the importance of chromatin accessibility as a dynamic and tunable feature that integrates environmental cues with intrinsic genetic programs. This duality enables microglia to swiftly adapt their functional repertoire in response to fluctuating pathological pressures, embodying a plasticity that is both a challenge and an opportunity for intervention. By cataloguing the accessible regulatory regions that govern microglial states, the study paves the way for designing epigenetic drugs aimed at restoring beneficial microglial functions.
The findings also carry profound implications for biomarker discovery. Chromatin accessibility signatures specific to disease-associated microglia could serve as sensitive indicators of disease stage or response to therapies. Such biomarkers, detectable in peripheral cells or cerebrospinal fluid, might offer minimally invasive diagnostics to track disease progression and assess treatment efficacy in real time. This precision could revolutionize clinical management of Alzheimer’s disease by enabling timely and targeted interventions.
Importantly, the research underscores the heterogeneity not only within microglial populations but also in the progression of Alzheimer’s pathology itself. The spatial gradients of chromatin accessibility mirror the spatial gradients of neuropathology, suggesting that effective therapies must consider both cellular and anatomical heterogeneity. This multi-dimensional approach challenges one-size-fits-all models and advocates for nuanced treatment strategies tailored to microglial niches within the brain.
In synthesizing these insights, Ardura-Fabregat and colleagues have charted new territory in the landscape of neurodegenerative disease research. By marrying spatial biology with epigenomics, they elucidate how microglia decode their microenvironment at the chromatin level to mount context-dependent responses. This conceptual framework not only enhances our comprehension of Alzheimer’s disease mechanisms but also redefines microglia as sophisticated interpreters of spatial and epigenetic information, orchestrating neuroinflammation with remarkable precision.
As the quest for effective Alzheimer’s therapies continues, this study stands as a testament to the power of integrating spatially resolved molecular technologies in unraveling complex brain diseases. It beckons researchers to probe deeper into the epigenetic underpinnings of microglial function and to harness this knowledge in designing next-generation interventions that could one day halt or reverse the devastating march of neurodegeneration.
Subject of Research: Microglial heterogeneity and chromatin accessibility in spatially defined brain regions in a mouse model of Alzheimer’s disease.
Article Title: Response of spatially defined microglia states with distinct chromatin accessibility in a mouse model of Alzheimer’s disease.
Article References:
Ardura-Fabregat, A., Bosch, L.F.P., Wogram, E. et al. Response of spatially defined microglia states with distinct chromatin accessibility in a mouse model of Alzheimer’s disease. Nat Neurosci (2025). https://doi.org/10.1038/s41593-025-02006-0
Tags: Alzheimer’s disease progression and microgliaanatomical niche effects on microglial activitychromatin architecture in brain immune cellsdeciphering microglial identity and functionepigenetic regulation of microglial cellsmicroglia dynamics in Alzheimer’s diseasemouse model studies of neurodegenerative diseasesneuroinflammatory responses in Alzheimer’sregional heterogeneity of microgliasingle-cell techniques in neurosciencespatial transcriptomics in neurodegenerationtransformative approaches in Alzheimer’s research