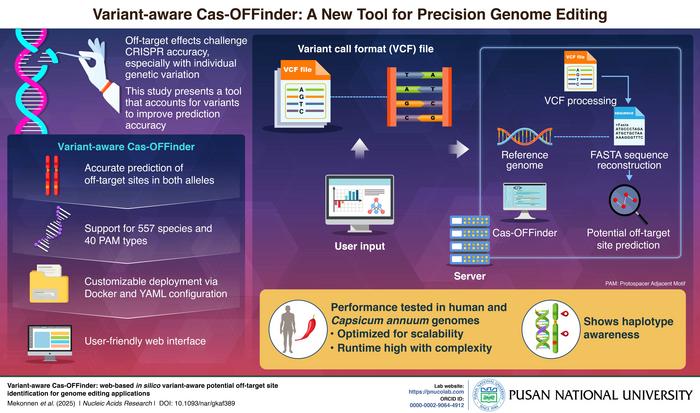
In recent years, the revolutionary gene-editing technology CRISPR-Cas9 has emerged as a powerful tool set to transform medicine, agriculture, and biological research. Despite its immense promise, one of the most persistent challenges facing CRISPR-based interventions is the accurate prediction and minimization of off-target effects—unintended alterations to the genome that can have deleterious consequences. These inadvertent edits arise partly due to the complexities and variabilities inherent in individual genomes, which differ extensively at the single nucleotide, insertion/deletion (indel), and larger structural levels. Traditionally, off-target prediction algorithms have relied heavily on standard reference genomes, which fail to capture this vital genetic diversity across individuals and alleles. Addressing this gap, a groundbreaking new web-based tool, Variant-aware Cas-OFFinder, has been introduced by a research team from Pusan National University in South Korea to significantly enhance off-target site identification by integrating personal genomic variation into the prediction process.
The pioneering work, spearheaded by Professor Jeongbin Park and co-first authors Abyot Melkamu Mekonnen and Kang Seong, introduces an innovative computational platform that acknowledges that CRISPR off-target activity is not a one-size-fits-all phenomenon but varies substantially based on allele-specific sequence context. This insight forms the foundation of Variant-aware Cas-OFFinder, which accepts phased single-sample Variant Call Format (VCF) files containing detailed genetic variant information. The tool reconstructs allele-specific genome sequences by incorporating single nucleotide polymorphisms (SNPs) along with insertions and deletions from the individual’s genotype data. This haplotype-level reconstruction enables the algorithm to perform off-target site predictions that reflect the unique genomic landscape of each allele, thereby offering unprecedented precision in identifying potential CRISPR editing risks.
While existing bioinformatics tools such as Cas-OFFinder and CRISPRitz provide rapid scanning of reference genomes for potential off-target sites, their reliance on canonical reference sequences inevitably overlooks individual-specific variants that may generate novel off-target loci or mask others. Variant-aware Cas-OFFinder transcends this limitation by enabling variant-aware scanning. By considering the full spectrum of small variants, the tool identifies off-target sites attributable specifically to insertions and deletions, which are often neglected in prior analyses. This significantly elevates the sensitivity and specificity of off-target prediction, setting a new standard for computational genome editing safety assessments.
.adsslot_3Ixh1TsgVU{width:728px !important;height:90px !important;}
@media(max-width:1199px){ .adsslot_3Ixh1TsgVU{width:468px !important;height:60px !important;}
}
@media(max-width:767px){ .adsslot_3Ixh1TsgVU{width:320px !important;height:50px !important;}
}
ADVERTISEMENT
Equipped with robust GPU acceleration compatibility, Variant-aware Cas-OFFinder supports resource-intensive haplotype-level analyses while maintaining practical computational efficiency. The tool currently accommodates genetic data from an impressive range of 557 species and supports 40 Protospacer Adjacent Motif (PAM) types, showcasing remarkable versatility across biological domains. This extensibility opens avenues for personalized genome editing applications not only in human health but also in agriculture and environmental sciences, where accurate off-target prediction tailored to diverse species and cultivars is vital.
Critically validating their tool, the Pusan National University team applied Variant-aware Cas-OFFinder to human and sweet pepper (Capsicum annuum) genomes, utilizing both public datasets and cultivar-specific sequencing information. In human samples, the analysis uncovered potential off-target sites on chromosome 10 that were absent from the standard human reference genome, highlighting the necessity of including personal genomic variation for reliable CRISPR design in clinical contexts. In the agricultural context, the tool identified allele-specific off-targets within sweet pepper cultivars, demonstrating how such haplotype-aware analyses can facilitate precision breeding strategies and accelerate the development of improved plant varieties with minimized genomic risks.
Fundamental to the philosophy of this project is the assertion from Prof. Park that genome editing must be as individualized as the very genetic material it seeks to modify. This resonates with emerging trends toward personalized medicine, where therapeutic interventions increasingly take into account patient-specific genetic landscapes. By fostering precise off-target prediction at the haplotype level, Variant-aware Cas-OFFinder offers a technological foundation for safer and more ethical clinical genome editing, reducing risks of unintended mutagenesis that could lead to oncogenic or other adverse outcomes.
Balancing computational complexity with predictive accuracy, the haplotype-level analyses implemented by Variant-aware Cas-OFFinder may introduce modest performance overheads compared to earlier tools limited to reference genomes. However, this trade-off is justified by the substantial increase in result fidelity and the capability to reveal personalized off-target profiles otherwise obscured in traditional analyses. This paradigm shift embraces the genomic intricacies of individuals rather than forcing them into a standardized mold.
The implications of this tool extend beyond academic research, heralding transformative possibilities in therapeutic genome editing. CRISPR therapies aiming to correct deleterious mutations in patients’ cells can now be refined to avoid off-targeting that may jeopardize patient safety. Similarly, agricultural biotechnologists can harness these insights to safely engineer crop genomes, tailoring modifications to specific cultivars’ genetic backgrounds and thus enhancing both efficacy and regulatory compliance.
As genome editing technologies continue to mature, the integration of variant-aware computational tools like Cas-OFFinder will be indispensable for minimizing off-target effects and realizing the full potential of precision medicine and sustainable agriculture. This innovation from Pusan National University marks a vital step towards the ultimate goal of personalized genome engineering that respects individual genetic uniqueness while maximizing safety and efficacy.
In summary, Variant-aware Cas-OFFinder represents a major advance in the computational genomics field by offering a haplotype-resolved, variant-informed approach to CRISPR off-target prediction. Its development addresses longstanding limitations of previous tools, builds a robust platform adaptable to hundreds of species, and delivers critical insights for both biomedical and agricultural genome editing applications. As the scientific community embraces such sophisticated tools, the promise of CRISPR as a truly precise genome engineer comes closer to fruition.
Subject of Research: Not applicable
Article Title: Variant-aware Cas-OFFinder: web-based in silico variant-aware potential off-target site identification for genome editing applications
News Publication Date: 8-May-2025
References: DOI: 10.1093/nar/gkaf389
Image Credits: Credit: Professor Jeongbin Park from Pusan National University, Korea
Keywords: CRISPRs, Genome editing, Computational biology, Bioinformatics, Genetic variation, Technology, Health and medicine, Plant genomes, Haplotypes
Tags: allele-specific CRISPR activitycomputational biology innovationsCRISPR gene editingCRISPR technology challengesgene-editing accuracy improvementsgenetic variant prediction toolsoff-target effects in CRISPRpersonalized genomic variationPusan National University researchsingle nucleotide variations in genomesVariant-aware Cas-OFFinderweb-based genetic tools