A groundbreaking study from Tokyo University of Science (TUS) has redefined our understanding of carbohydrate-active enzymes by uncovering entirely new families of β-1,2-glucanases—enzymes that play a critical role in the breakdown of complex bacterial carbohydrates. Published in the prestigious journal Protein Science on May 24, 2025, this research sheds light on the complex enzymology behind β-1,2-glucan degradation and introduces an innovative phylogenetic framework known as the “SGL clan,” expanding the horizons of molecular enzymology and carbohydrate metabolism.
Carbohydrates are fundamental biomolecules indispensable for life, serving both as crucial energy reservoirs and structural components across all domains of life. Among sugars, β-1,2-glucans occupy a unique niche. These glucose-based polysaccharides are predominantly bacterial in origin and are involved in myriad physiological functions including bacterial pathogenicity and adaptability to environmental pressures. Despite their significance, studying β-1,2-glucans has been challenging due to their relative scarcity and highly intricate structural arrangements, which stand in contrast to more commonly studied polysaccharides like cellulose.
The TUS research team, led by Associate Professor Masahiro Nakajima, embarked on an ambitious project to uncover new glycoside hydrolase (GH) enzymes capable of degrading these elusive β-1,2-glucans. Their approach combined in-depth sequence analyses, biochemical characterization, structural elucidation, and phylogenetic investigations, targeting a cluster of previously unclassified GH enzymes suspected of being related to known β-1,2-glucanases within GH families 144 and 162. This multifaceted methodological framework allowed the team to identify new enzyme families exhibiting β-1,2-glucanase activity, marking a considerable leap forward in carbohydrate enzymology.
.adsslot_OyC06NdQT8{width:728px !important;height:90px !important;}
@media(max-width:1199px){ .adsslot_OyC06NdQT8{width:468px !important;height:60px !important;}
}
@media(max-width:767px){ .adsslot_OyC06NdQT8{width:320px !important;height:50px !important;}
}
ADVERTISEMENT
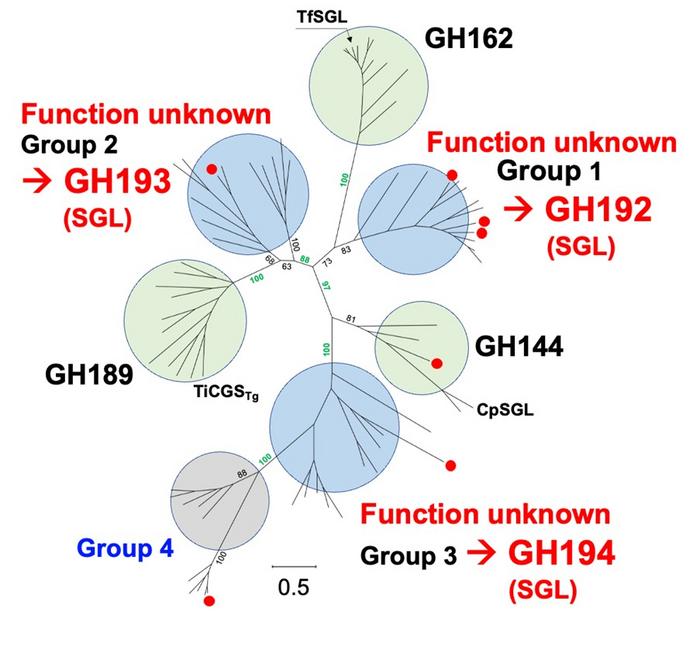
Central to their findings was the identification of four novel glycoside hydrolase families, three of which demonstrated enzymatic activity consistent with β-1,2-glucan degradation. Despite exhibiting as low as 16–20% amino acid sequence similarity among themselves, these enzymes were unified by shared structural motifs, most notably the (α/α)6-barrel fold, a feature previously identified in certain glycosidases. Furthermore, these new enzymes exhibited an identical anomer-inverting hydrolytic mechanism — a fundamental catalytic strategy whereby the stereochemistry at the anomeric carbon is inverted during glycosidic bond cleavage.
Such structural and mechanistic congruencies led the research team to propose the formation of a new classification, termed the “SGL clan,” encompassing three freshly characterized families GH192, GH193, GH194 alongside the established GH144 and GH162 families. Intriguingly, the GH189 family, which employs an anomer-retaining catalytic mechanism, was also included within this clan owing to phylogenetic relations and functional characteristics. This clan-based classification not only groups sharing evolutionary traits but also emphasizes the remarkable molecular diversity underscored by their catalytic features.
One of the most compelling aspects of this study lies in the molecular evolution of these enzymes. The researchers demonstrated that the irregular distribution of catalytic reaction mechanisms across the SGL clan correlates phylogenetically to variations in the positions of key catalytic residues. This suggests a unique evolutionary trajectory, through which substrate specificity and enzymatic mechanisms diversified from common ancestors. Despite their functions converging on β-1,2-glucan cleavage, these enzymes share only three strictly conserved amino acid residues—E239, Y367, and F286—which the authors identify as the defining molecular signature of the SGL clan.
This focused conservation amidst widespread sequence divergence highlights an evolutionary strategy that balances functional conservation with sequence diversification. It also provides a powerful molecular marker to identify and classify new β-1,2-glucanases within this clan, potentially accelerating the discovery of additional enzymes with related functions but novel sequences.
The study’s implications extend far beyond taxonomy and enzymology. Glycans are notoriously complex, and their structural diversity makes their enzymatic synthesis and degradation a technically demanding endeavor. By unraveling the molecular underpinnings of β-1,2-glucanase activity and expanding the repertoire of enzymes capable of mediating these reactions, the research opens new pathways for engineering synthetic and degradative enzymes. Such progress has transformative potential for diverse applications, including the tailored synthesis of oligosaccharides for therapeutic use, improved understanding of bacterial virulence mechanisms, and novel biotechnological tools for sustainable biofuel production.
Importantly, this research exemplifies the dynamic interplay between glycan synthesis and degradation. As Dr. Nakajima notes, the synergistic exploration of both enzymatic synthesis and degradation significantly expands our comprehension of glycan biology. Not only do these enzymes have roles in catabolism, but they also bear the potential for engineered synthetic applications where modifying enzyme specificity and function could yield custom-designed oligosaccharides — molecules critical for pharmaceuticals, diagnostics, and beyond.
The interdisciplinary collaboration that fueled these discoveries deserves recognition. Dr. Nakajima’s leadership, supported by contributions from former doctoral student Dr. Sei Motouchi, Associate Professor Hiroyuki Nakai from Niigata University, and Dr. Kaito Kobayashi from the National Institute of Advanced Industrial Science and Technology, represented a collaborative synergy that leveraged expertise spanning structural biology, enzymology, and phylogenetics. Their integrated approach exemplifies how cross-institutional collaboration can accelerate innovation in glycobiology research.
Furthermore, the revelation that the evolutionary distribution of enzyme catalytic mechanisms is tightly linked to the spatial organization of essential residues adds a new layer of understanding to how enzymatic function evolves. This knowledge facilitates rational enzyme engineering, allowing scientists to anticipate how modifications in residue positioning might shift reaction stereochemistry or substrate specificity, paving the way for customized enzyme design.
From a broader perspective, uncovering the molecular evolution pathways of these enzymes enriches the ever-growing catalogue of carbohydrate-active enzymes (CAZymes), a group crucial to biotechnology and life sciences. This sophisticated understanding of SGL clan enzymes highlights the evolutionary adaptability of bacteria and indicates potential targets for antibiotic development, given the role of β-1,2-glucans in bacterial physiology and pathogenicity.
In conclusion, the identification and characterization of new glycoside hydrolase families within the SGL clan deeply enrich our understanding of carbohydrate enzymology. This study not only expands the known diversity of β-1,2-glucan-degrading enzymes but also offers fresh mechanistic insights and evolutionary context that could transform future research and industrial applications. The newly proposed clan classification harmonizes complex biochemical and phylogenetic data into a cohesive framework, setting the stage for novel explorations in synthetic carbohydrate chemistry, microbial biology, and enzyme engineering.
As advances continue, the possibility of converting naturally degradative enzymes into synthetic catalysts to design novel oligosaccharides may soon become a practical reality. This would mark a paradigm shift in carbohydrate science, with wide-reaching implications for medicine, agriculture, and renewable bio-based industries. The discovery of the SGL clan thus stands as a testament to the power of interdisciplinary research to unveil nature’s molecular innovations and inspire next-generation biotechnologies.
Subject of Research: Not applicable
Article Title: New glycoside hydrolase families of β-1,2-glucanases
News Publication Date: 24-May-2025
Web References:
https://doi.org/10.1002/PRO.70147
References:
Nakajima, M., Tanaka, N., Motouchi, S., Kobayashi, K., Shimizu, H., Abe, K., Hosoyamada, N., Abara, N., Morimoto, N., Hiramoto, N., Nakata, R., Takashima, A., Hosoki, M., Suzuki, S., Shikano, K., Fujimaru, T., Imagawa, S., Kawadai, Y., Wang, Z., Kitano, Y., Nihira, T., Nakai, H., Taguchi, H. (2025). New glycoside hydrolase families of β-1,2-glucanases. Protein Science, 34(6). https://doi.org/10.1002/PRO.70147
Image Credits: Associate Professor Masahiro Nakajima, Tokyo University of Science, Japan
Keywords: β-1,2-glucanase, glycoside hydrolase, enzyme evolution, carbohydrate metabolism, glycan degradation, SGL clan, enzyme classification, molecular enzymology, phylogenetic analysis, structural biology, enzyme engineering, carbohydrate-active enzymes
Tags: 2-glucanasesbacterial carbohydrate degradationbacterial pathogenicity and adaptabilitybiochemical characterization and analysiscarbohydrate metabolism advancementscarbohydrate-active enzymescomplex enzymology studiesglycoside hydrolase enzymesnovel enzyme familiespolysaccharide structural analysisSGL clan phylogenetic frameworkTokyo University of Science researchβ-1