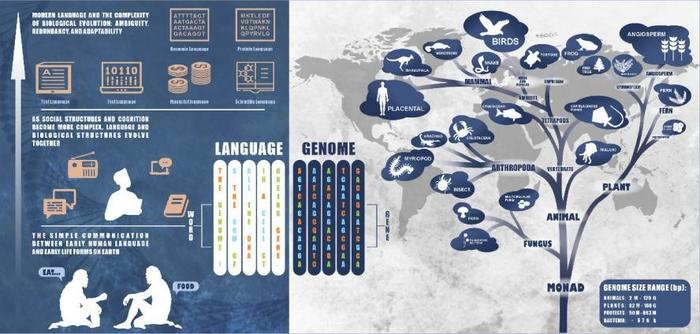
In a groundbreaking study published in the prestigious journal Tropical Plants, a team of researchers from Hainan University has made significant strides in the integration of artificial intelligence (AI) within plant genomics. By utilizing large language models (LLMs), this research opens new avenues for decoding complex genetic information, akin to interpreting a new language. The implications are profound, especially regarding agricultural innovation, biodiversity conservation, and the global challenge of food security.
For years, the field of plant genomics has faced substantial challenges due to the immense and intricate nature of genetic data. Traditional approaches to analyzing genomic information often falter in the face of vast datasets that include numerous species and their associated genomic variations. Moreover, classical machine learning techniques struggle with the specificity and resolution needed in this domain, especially when annotated data is scarce. The introduction of LLMs introduces a paradigm shift in how scientists approach plant genomic analysis, enabling them to leverage the structural and functional parallels between genetic sequences and human language.
This research specifically addresses the need to adapt LLMs to comprehend the unique nuances of plant genomes. Unlike human languages, which adhere to grammatical and syntactical rules, plant genomes operate under a set of biological rules that govern gene expression and regulation. The researchers detail a method that involves training LLMs on extensive datasets of plant genomic information, allowing these models to recognize patterns and make predictions regarding gene functions and regulatory elements.
.adsslot_Jdhgyb1AC0{width:728px !important;height:90px !important;}
@media(max-width:1199px){ .adsslot_Jdhgyb1AC0{width:468px !important;height:60px !important;}
}
@media(max-width:767px){ .adsslot_Jdhgyb1AC0{width:320px !important;height:50px !important;}
}
ADVERTISEMENT
Through their innovative work, the authors highlight the training process for these models, which involves two critical phases: pre-training and fine-tuning. In the pre-training stage, LLMs consume vast amounts of unannotated plant genomic data, equipping them to identify underlying similarities between different genomic sequences. Following this, the fine-tuning phase utilizes specific annotated datasets to refine the model’s predictive capabilities, ensuring that it can accurately interpret the biological functions encoded within the genomic sequences.
One of the remarkable findings of this study is the successful application of different LLM architectures tailored for plant genomics. The researchers explored encoder-only models like DNABERT, which focus on parsing an input sequence to create a comprehensive representation; decoder-only models such as DNAGPT, which generate outputs based on learned patterns; and encoder-decoder models like ENBED, which synthesize elements of both approaches. Each model presents unique strengths in handling various aspects of genomic data, from the identification of enhancers and promoters to the prediction of gene expression patterns.
The findings also introduce plant-specific models like AgroNT and FloraBERT, which have demonstrated enhanced performance in annotating plant genomes compared to their more generalized counterparts. By focusing on the linguistic characteristics of DNA sequences, these models exhibit superior ability to unravel the complexities of gene regulation and enable the application of genomic information in practical agricultural contexts.
Despite this progress, the study acknowledges significant gaps that remain in existing LLM architectures. A majority of current models are predominantly trained on datasets derived from animal or microbial organisms, which frequently lack the comprehensive genomic annotations necessary for effective application in plant species. The authors advocate for further development of plant-focused LLMs that can incorporate diverse genomic datasets, especially from lesser-studied species like tropical plants, to achieve a more representative understanding of plant biology and genetics.
In an era where climate change and population growth present imminent threats to global food security, the insights gained from this research are immensely valuable. By harnessing the potential of AI and LLMs in plant genomics, researchers can pave the way for accelerated crop improvement strategies, better adaptation of plant species to changing environmental conditions, and ultimately enhanced biodiversity conservation efforts.
In summary, the research underscores the transformative potential of integrating AI-driven methodologies within the field of plant genomics. By bridging the gap between computational linguistics and genetic analysis, researchers are poised to revolutionize our understanding of plant biology. This not only promises to enhance agricultural productivity but also aims to foster sustainable practices that address the pressing challenges of the 21st century.
As the field advances, future endeavors will likely focus on refining LLM architectures, expanding training datasets to include a broader array of plant species, and investigating real-world agricultural applications. By continuing to push the boundaries of what is possible at the intersection of artificial intelligence and genomics, we stand to unlock a wealth of knowledge that could improve our critical relationship with the natural world.
This pivotal study sets the stage for a new era in plant genomic research, one where AI is not just an auxiliary tool but a central player in unraveling the complexities of life at the genetic level.
Subject of Research: Plant Bio-Genomics
Article Title: Artificial Intelligence-Driven Plant Bio-Genomics Research: A New Era
News Publication Date: 14-Apr-2025
Web References: Tropical Plants
References: 10.48130/tp-0025-0008
Image Credits: The authors
Keywords
Research methods, applied sciences and engineering.
Tags: advanced machine learning for plant geneticsagricultural innovation through AIAI in plant genomicsartificial intelligence and biodiversitydecoding plant DNA with AIfood security and genomicsgenomic data analysis challengesintegration of AI and agriculturelarge language models for agricultureLLMs revolutionizing genetic researchnovel approaches to genomic informationunderstanding plant genome complexity