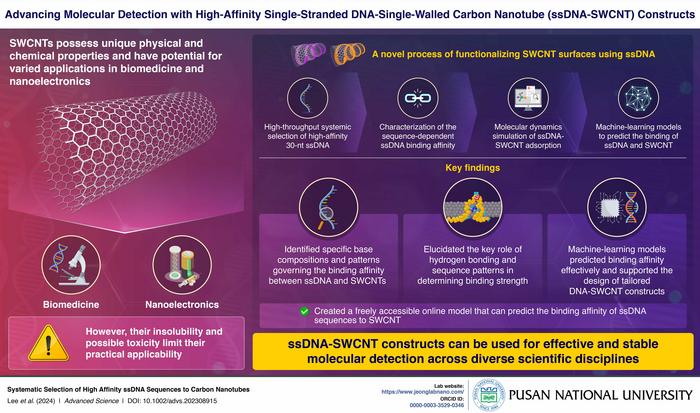
Single-walled carbon nanotubes (SWCNTs) have emerged as promising candidates for applications in biotechnology and nanoelectronics due to their exceptional physical and chemical properties. Despite their potential, challenges like insolubility and toxicity have hindered their widespread use. Prior studies have been investigating diverse strategies to functionalize and modify the surfaces of SWCNTs to overcome these challenges.
Credit: Professor Sanghwa Jeong from Pusan National University, Korea
Single-walled carbon nanotubes (SWCNTs) have emerged as promising candidates for applications in biotechnology and nanoelectronics due to their exceptional physical and chemical properties. Despite their potential, challenges like insolubility and toxicity have hindered their widespread use. Prior studies have been investigating diverse strategies to functionalize and modify the surfaces of SWCNTs to overcome these challenges.
In a recent study, researchers from the Pusan National University led by Professor Sanghwa Jeong, Assistant Professor in the School of Biomedical Convergence Engineering, have attempted to fill this gap. This study has gone beyond conventional techniques by employing high-throughput screening methods to elucidate the relationship between DNA sequences and their binding affinity to carbon nanotubes. It focused on optimizing the binding affinity and stability of these constructs through advanced sequence design and molecular dynamics simulations. This recent study was published in the journal of Advanced Science on 25th June 2024. Discussing the background of their study Dr. Jeong explains, “Researchers have been exploring various strategies to engineer SWCNT surfaces to overcome the challenges of limited applications owing to insolubility and potential toxicity. One promising approach is the use of single-stranded DNA (ssDNA) as a wrapping surfactant for SWCNTs.”
The researchers employed a rigorous methodology to ensure precise characterization and optimization of single-stranded DNA (ssDNA)-SWCNT complexes. Initially, a diverse random 30-nucleotide (nt) ssDNA library underwent iterative rounds of screening to identify high-affinity sequences.
Computational modeling, particularly molecular dynamics simulations, provided insights into the structural dynamics of the SWCNT constructs. Furthermore, the researchers used several machine-learning models to understand the pattern of sequences that affect binding affinity. They have successfully created a freely accessible online service that predicts the binding affinity of ssDNA sequences to SWCNTs. These integrated approaches not only validated the experimental findings but also guided the design of high-performance ssDNA-SWCNT constructs.
The findings revealed significant advancements in the stability and functionality of ssDNA-SWCNT complexes. High-affinity 30-nt ssDNA sequences, rich in adenine and cytosine, exhibited superior binding strength, validated through surfactant displacement experiments. Molecular dynamics simulations highlighted the formation of stable intramolecular hydrogen bonds near the SWCNT surface, underscoring their enhanced structural integrity. The machine-learning models effectively predicted the binding affinities of ssDNA sequences, further supporting the design of the tailored ssDNA-SWCNT constructs.
Moreover, the study demonstrated notable improvements in the resistance of these complexes to enzymatic degradation compared to free ssDNA, making them highly suitable for long-term biological applications.
In conclusion, the development of high-affinity ssDNA-SWCNT constructs marks a significant advancement in nanobiotechnology. The exceptional characteristics of ssDNA-SWCNTs make them ideal candidates for cell or tissue-specific drug delivery systems as well as the development of high-performance nano electronic devices.
Dr. Jeong concludes, “Our study not only makes a substantial contribution to our understanding of the interplay between ssDNA and SWCNTs but also offers practical avenues for harnessing these interactions in a wide range of advanced technologies. In the future, developing nanomaterials and devices with enhanced stability will show promise in driving innovation in nanoelectronics and biotechnology.”
***
Reference
DOI: https://doi.org/10.1002/advs.202308915
Website: https://www.jeonglabnano.com/
Orchid Id: 0000-0003-3529-0346
About the institute
Pusan National University, located in Busan, South Korea, was founded in 1946 and is now the No. 1 national university of South Korea in research and educational competency. The multi-campus university also has other smaller campuses in Yangsan, Miryang, and Ami. The university prides itself on the principles of truth, freedom, and service, and has approximately 30,000 students, 1200 professors, and 750 faculty members. The university is composed of 14 colleges (schools) and one independent division, with 103 departments in all.
Website: https://www.pusan.ac.kr/eng/Main.do
About the author
Prof. Sanghwa Jeong is an Assistant Professor in the School of Biomedical Convergence Engineering at Pusan National University. His group is developing the novel optical nanoprobe for biological signaling, especially neurochemical signaling via highly interdisciplinary research techniques including nanoscience, chemistry, bioengineering, and data science. They aim to establish the dynamic in vivo neurochemical imaging toolkit, which will unveil the hidden stages of brain functions and elicit the discovery of promising cures and therapies for various neurological disorders.
Lab website: https://www.jeonglabnano.com/
ORCID ID: 0000-0003-3529-0346
Journal
Advanced Science
DOI
10.1002/advs.202308915
Method of Research
Experimental study
Subject of Research
Not applicable
Article Title
Systematic Selection of High Affinity ssDNA Sequences to Carbon Nanotubes
Article Publication Date
25-Jun-2024
COI Statement
The authors declare that they have no competing interests.




