This study is led by Prof. Xianqun Fan (Department of Ophthalmology, Shanghai Jiao Tong University School of Medicine Affiliated Ninth People’s Hospital).
Credit: ©Science China Press
This study is led by Prof. Xianqun Fan (Department of Ophthalmology, Shanghai Jiao Tong University School of Medicine Affiliated Ninth People’s Hospital).
Gene editing ushers in a new era of disease treatment since many genetic diseases are caused by base-pair mutations in genomic DNA. With the rapid development of genome editing technology, novel editing tools such as base editing and prime editing have attracted public attention, heralding a great leap forward in this field.
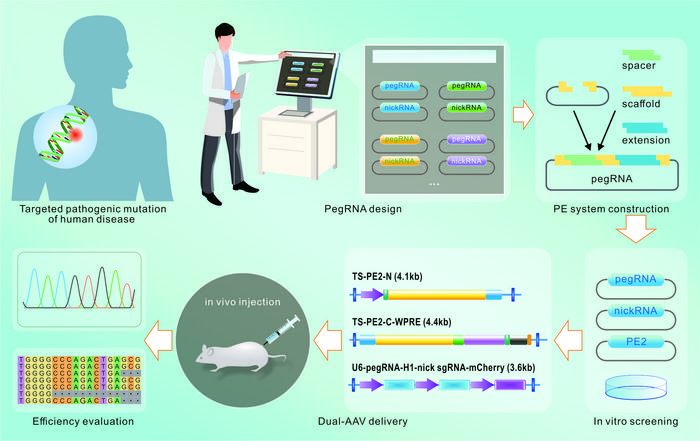
Prime editing (PE) was proposed by David Liu’s team in 2019, which is characterized by no need for double-strand breaks (DSBs) or homology sequence templates with variable application scenarios, including point mutations as well as insertions or deletions. The PE system is composed of two parts: prime editors (PEs) and prime editing guide RNA (pegRNA). Prime editors are a kind of fusion proteins between a Streptococcus pyogenes Cas9 (SpCas9) H840A nickase mutant and an engineered reverse transcriptase (RT) domain. The pegRNA functions as both sgRNA and donor template for the desired alteration. It contains a spacer sequence, a scaffold, and a 3’extension that is formed of primer binding sequence (PBS) and RT template (RTT). The spacer sequence will specify the target site. After nicking the PAM-containing DNA strand by the Cas9 nickase domain, the resulting 3’ end will hybridize with the PBS and serve as a DNA primer for reverse transcription by the RT domain.
When delivered into a cell, the PEs are targeted to the editing site and nick the single PAM-containing DNA strand, resulting in a release of the 3ʹ DNA end that can hybridize to the pegRNA extension part. Afterward, the reverse transcriptase performs polymerization of edited DNA from the RT template of pegRNA into the target site, which generates either a 3’ flap with an edit or a 5’ flap without an edit. The preference for 5’ flap excision and 3’ flap ligation finally creates heteroduplex DNA containing one edited strand and one unedited strand. DNA repair or replication copies the edit to the complementary strand and permanently installs the desired edit.
The PE technology has grown and rapidly progressed in the past four years, with versatile advancements in their architecture to increase editing efficiency, targetability, and specificity, including novel pegRNA design, PE modification, and delivery improvement. Additionally, despite relatively recent inception, PE has been extensively applied to correct pathologic mutations from genetic diseases both in vitro and in vivo, which has great potential for moving the field of gene therapy forward from bench to bedside.
In this review, the authors in detail describe the developmental history of prime editing, the newest evolution of CRISPR‒Cas-based technologies. Websites for pegRNA design have been listed, including pegFinder, pegIT, PrimeDesign, and so on. A variety of sequencing methods for off-target prediction are also elaborated upon. Then, the application of PE in human disease models is briefly summarized in the following section, ranging from eye diseases to liver hereditary disorders. With higher editing efficiency and fewer byproducts than traditional editing tools, prime editing holds great promise as a therapeutic strategy for human diseases.
In conclusion, PE is a tremendous milestone in the development of a universal method for genome editing. Its clinical adaptation toward the precise correction of known pathogenic mutations in human diseases may catapult the next frontier of personalized treatments.
Journal
Science Bulletin
DOI
10.1016/j.scib.2023.11.015