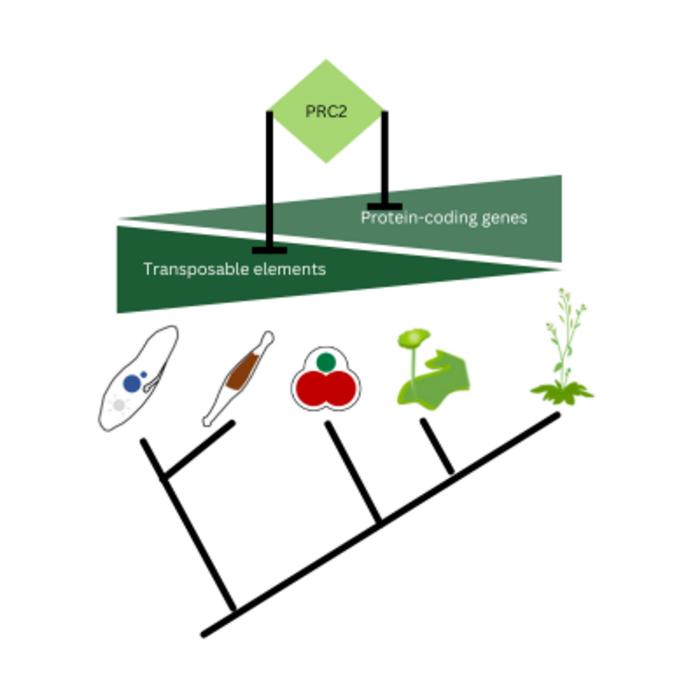
Polycomb repressive complex 2 (PRC2) was discovered decades ago in Drosophila, where it was found to be a key controller of developmental genes. Further analyses showed that PRC2 modifies chromatin and silences target gene expression. However, the ancestral function of PRC2 – as functioning primarily to control genes during development – was called into question when researchers discovered that PRC2 also plays a role in unicellular species, in which no development takes place. A first hint at PRC2’s original role came from studies in red algae, which found PRC2 left its methylation mark on transposons – jumping genes that move around the genome. Frederic Berger and his research group at the Gregor Mendel Institute of Molecular Plant Biology (GMI) decided to follow this cue and, in an international collaboration with researchers at Freie Universität Berlin, University of Cambridge, Nantes University, the National Institute of Genetics (Japan) and Monash University, investigated how PRC2 acts in a range of eukaryotes.
Credit: (c) GMI
Polycomb repressive complex 2 (PRC2) was discovered decades ago in Drosophila, where it was found to be a key controller of developmental genes. Further analyses showed that PRC2 modifies chromatin and silences target gene expression. However, the ancestral function of PRC2 – as functioning primarily to control genes during development – was called into question when researchers discovered that PRC2 also plays a role in unicellular species, in which no development takes place. A first hint at PRC2’s original role came from studies in red algae, which found PRC2 left its methylation mark on transposons – jumping genes that move around the genome. Frederic Berger and his research group at the Gregor Mendel Institute of Molecular Plant Biology (GMI) decided to follow this cue and, in an international collaboration with researchers at Freie Universität Berlin, University of Cambridge, Nantes University, the National Institute of Genetics (Japan) and Monash University, investigated how PRC2 acts in a range of eukaryotes.
To understand PRC2’s role in the ancestors of eukaryotes, the researchers studied the genomes of three widely distant lineages of eukaryotes: plants, SAR and ophistokonts, the lineage to which humans and fungi belong. All of these eukaryotes contain transposons, mobile genetic elements that can excise themselves from DNA and insert at novel locations – a potential threat to genome stability, hence why transposons have to be silenced. In mutant diatoms, bryophytes and red algae the removal of PRC2 activity caused the loss of silencing of transposable elements.
“These findings show that PRC2 represses transposable elements in these distant lineages, which defines this as a function that arose in the ancestor all three lineages. PRC2’s origin was likely to primarily defend the genome from invasion by transposons. This ancestral function is a profound change in paradigm”, says Frederic Berger, group leader at GMI and the study’s corresponding author.
Over the course of evolution, however, the function of PRC2 gradually shifted from repressing transposons to repressing protein-coding genes, its originally described role. In land plants, the researchers find “fossil TEs” to still be targets for PRC2, which then silences genes in the neighborhood. “In flowering plants, like Arabidopsis, which evolved more recently, remnants of TEs are still present and recruit PRC2 to silence nearby genes”, explains Tetsuya Hisanaga, a postdoctoral researcher in the Berger group and the study’s first author. “We hypothesize that in Arabidopsis, some TEs have been domesticated, becoming elements involved in modulating protein-coding genes.”
Journal
Current Biology
DOI
10.1016/j.cub.2023.08.073
Method of Research
Experimental study
Subject of Research
Not applicable
Article Title
The Polycomb Repressive Complex 2 deposits H3K27me3 and represses transposable elements in a broad range of eukaryotes.
Article Publication Date
21-Sep-2023
COI Statement
The authors declare no competing interests.




