Lower respiratory tract infection (LRTI), which includes conditions such as pneumonia, has long been the leading cause of death from communicable agents and a leading cause of death in children worldwide. But despite its prevalence, LRTI is tricky for doctors to treat effectively because the current diagnostic approach often fails to conclusively determine whether an infection is present at all, and if so, what pathogen is causing it.
Credit: Mick, et al.
Lower respiratory tract infection (LRTI), which includes conditions such as pneumonia, has long been the leading cause of death from communicable agents and a leading cause of death in children worldwide. But despite its prevalence, LRTI is tricky for doctors to treat effectively because the current diagnostic approach often fails to conclusively determine whether an infection is present at all, and if so, what pathogen is causing it.
Now, in a study published April 3, 2023 in The Journal of Clinical Investigation, a team led by researchers at the Chan Zuckerberg Biohub San Francisco (CZ Biohub SF), UC San Francisco (UCSF), the University of Colorado Anschutz Medical Campus, and the University of Arkansas for Medical Sciences (UAMS)/Arkansas Children’s Research Institute (ACRI) describe a new method for LRTI diagnosis in children with severe respiratory failure. The method applies machine learning to metagenomic sequencing data obtained from lung fluid of intubated children to diagnose pediatric LRTI and to identify its cause with very high accuracy, far exceeding current techniques.
Shots in the dark
LRTI may be caused by a wide variety of bacterial, viral, or fungal pathogens, but the symptoms of infection appear clinically similar no matter the culprit and are even difficult to distinguish from noninfectious respiratory conditions. Because of the limitations of existing diagnostics, doctors often work under a “fog of war” when it comes to developing an effective treatment plan, said Eran Mick, a bioinformatics scientist at CZ Biohub SF and UCSF and one of the study’s lead authors.
Current diagnostics often rely on growing bacteria from lung-fluid samples in culture, which is time-consuming and doesn’t always correctly identify the bacterial species responsible for the problem. The tests often generate false-negative results or detect incidental microbes that aren’t actually causing an illness.
“In over half of all cases, the actual microbe causing the infection is not identified,” said corresponding author Chaz Langelier, a CZ Biohub SF Investigator and associate professor of medicine in UCSF’s Division of Infectious Diseases. “What results is that the treatments given are not necessarily directed at what’s causing the problem.”
Instead, doctors may prescribe a cocktail of broad-spectrum antibiotics in an effort to stop the suspected infection, which contributes to the emergence of bacteria with antibiotic resistance. This practice can also lead to adverse patient outcomes like kidney injury or infection by other harmful bacteria, such as Clostridioides difficile. And it may even be entirely ineffective against the causal pathogen, as in the case of viral infections.
“While viruses cause the majority of LRTIs in young children, we know that children with severe LRTI requiring mechanical ventilation support often have co-infections of a virus and bacteria,” said co–senior author Peter Mourani, professor of pediatrics in Critical Care Medicine at UAMS and ACRI president. “So even if a virus is detected using commonly employed PCR testing, clinicians often feel compelled to treat for a possible bacterial infection, including when clinical cultures are negative. Most children receive antibiotics prior to specimen collection which may then contribute to false-negative culture results.”
“Adding to the challenges with diagnosing LRTI is that the true biology of lower respiratory infections is not just about a pathogenic microbe,” Langelier said. “It’s really a dynamic interplay between the pathogen causing the infection, the patient’s immune response, and the lung’s normal microbiome.”
A more holistic approach
To better diagnose and treat LRTI in children, the researchers developed a new method that takes into account not only the presence of potential pathogens in the lung but also the patient’s immune response, which can indicate whether the body is actually fighting off an infection.
The method relies on metagenomic RNA sequencing, which simultaneously reports on the patient’s gene expression and on any microbes present, all from a single sample of lung fluid. Metagenomics captures all the genetic sequences present in a sample – whether from the patient, bacteria, or viruses. Those sequences can then be computationally compared to reference databases to quantify gene expression and the abundance of microbes.
The researchers then applied machine learning algorithms to the metagenomic data to identify the joint patterns of gene expression and microbe abundance that distinguish actual LRTI from the unharmful presence of microbes that are an inherent part of the lung microbiome.
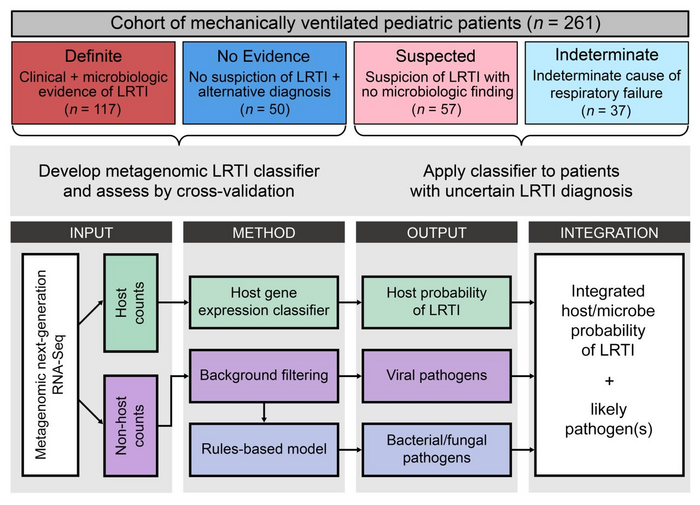
The researchers generated and studied metagenomic data from a cohort of 261 children with acute respiratory failure who had been admitted to one of eight children’s hospitals in the U.S. To properly train the machine learning algorithms, a team of clinicians rigorously adjudicated whether each of the children had LRTI, a clear noninfectious cause of respiratory failure, or an uncertain diagnosis.
Data from patients characterized as definitely suffering from LRTI and from patients having no evidence of LRTI were used to develop and then test the metagenomic diagnostic. In addition to making a diagnosis, the method also nominates the likeliest causal pathogen for the patients that are classified as infected.
The method proved to be incredibly accurate. “It worked better than expected,” Langelier said. “Our ‘area under the curve,’ which is essentially a measure of diagnostic test performance, was above 0.98, on a scale of 0 to 1. That’s about as good as one can get with a diagnostic test and is actually better than a similar test that we developed in adults.”
The concept for this work stemmed from previous research by Langelier, Katrina Kalantar of the Chan Zuckerberg Initiative (CZI), UCSF Professor and SF Biohub President Joe DeRisi, and their colleagues, in which they used metagenomics to effectively diagnose lower respiratory tract infections in critically ill adult patients.
A diagnostic for the 21st century
“This test would have a big impact clinically,” said Alexandra Tsitsiklis, a lead author of the study and a former member of Langelier’s lab now at New York City–based Immunai. “One of the key advantages of a metagenomic diagnostic is the ability to provide an answer in the patients with an uncertain clinical diagnosis.” Indeed, the method was able to diagnose LRTI and identify probable pathogens for many of the patients who didn’t fall into one of the two definitive clinical diagnosis categories used to train the machine-learning model.
In addition, Tsitsiklis said, a test like this could be used to rule out a bacterial infection, so that antibiotics could be avoided for these patients specifically, adding, “This is one of the next steps that would make it even more useful clinically.”
The study was also co-led by Jack Kamm, who was a member of CZ Biohub SF’s Data Science team at the time. Other coauthors include Katrina Kalantar from CZI, Norma Neff and Angela Detweiler of CZ Biohub SF’s Genomics Platform, and several researchers from the University of Colorado; see the paper for a complete list of authors.
The researchers are hopeful that following further validation, their diagnostic method will become commonplace in hospital settings. “The way that infectious disease diagnosis works is very primitive. It’s the same as it has been for decades and doctors aren’t happy with the tools they have. So we hope this is something we can keep working on to get to the point where it’s implemented in the hospital,” said Mick. “We are trying to set the stage for bringing hospitals into the 21st century.”
# # #
About the Chan Zuckerberg Biohub San Francisco
The Chan Zuckerberg Biohub San Francisco, part of the Chan Zuckerberg Biohub Network, is a nonprofit biomedical research center founded in 2016. CZ Biohub SF’s researchers, engineers, and data scientists, in collaboration with colleagues at our partner universities Stanford University; the University of California, Berkeley; and the University of California, San Francisco, seek to understand the fundamental mechanisms underlying disease and develop new technologies that will lead to actionable diagnostics and effective therapies. Learn more at czbiohub.org/sf.
Journal
Journal of Clinical Investigation
DOI
10.1172/JCI165904