CycPeptMPDB, a novel database—created by Tokyo Tech researchers—focused on the membrane permeability of cyclic peptides, could accelerate the development of drugs based on these promising compounds. This database was created by gathering published information on thousands of cyclic peptides and organizing it neatly in an online-accessible platform. Thanks to its search and visualization capabilities, CycPeptMPDB could pave the way to new computational and machine learning methods for screening and designing drugs with cyclic peptides.
Credit: Professor Yutaka Akiyama
CycPeptMPDB, a novel database—created by Tokyo Tech researchers—focused on the membrane permeability of cyclic peptides, could accelerate the development of drugs based on these promising compounds. This database was created by gathering published information on thousands of cyclic peptides and organizing it neatly in an online-accessible platform. Thanks to its search and visualization capabilities, CycPeptMPDB could pave the way to new computational and machine learning methods for screening and designing drugs with cyclic peptides.
One of the greatest challenges in modern drug design is to find compounds that satisfy somewhat contradictory requirements—they need to be small enough to permeate human cell membranes, while being large enough to target various protein surfaces and protein–protein interactions. This is a fine balance to achieve—if the compounds are too large, they may not pass through the cell membrane, and their bioavailability would be affected; if they are too small, they would not retain high specificity to the target protein (or proteins). Scientists estimate that over 80% of all known proteins associated with diseases cannot be targeted by conventional small-molecule drugs or antibody-based drugs. That is why, in recent years, cyclic peptides have become a very active research area. In principle, these compounds can achieve the fine balance required of modern drugs.
A cyclic peptide is a type of organic molecule that consists of amino acids linked together in a circular or lariat shape. What makes them particularly attractive is that they can target intracellular protein–protein interactions, which have been considered “undruggable” for decades. Moreover, cyclic peptides are inexpensive to synthesize compared to antibody-based drugs, prompting many pharmaceutical companies to conduct extensive research on these compounds.
However, one of the biggest hurdles to overcome in cyclic peptide research is that their membrane permeability—which controls their bioavailability and efficiency as drugs—is low in general, and the mechanisms behind this are not completely understood. Thus, during drug design, it is difficult for researchers to select candidate peptides that are likely to make it through the cell membrane. On top of this, there are currently no openly accessible databases documenting the membrane permeabilities of known cyclic peptides.
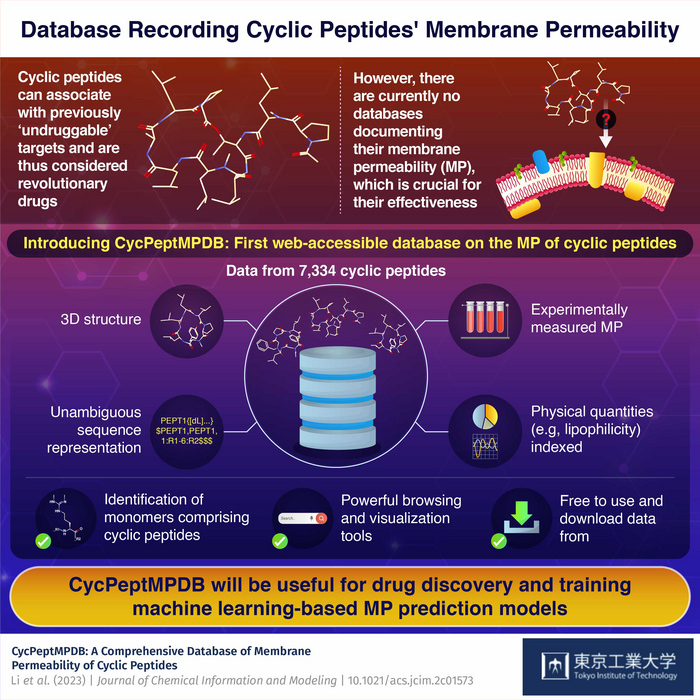
Against this backdrop, a team of researchers from Tokyo Institute of Technology (Tokyo Tech), Japan, including Professor Yutaka Akiyama, decided to take a step towards making cyclic peptide research easier for everyone. As explained in their latest paper published in the Journal of Chemical Information and Modeling, the team created an online database called CycPeptMPDB that contains information on thousands of cyclic peptides, including their membrane permeability.
To build the database, they gathered data from previously published papers and pharmaceutical patents. After inspecting over 40 publications, they collected information on 7,334 cyclic peptides with widely different chemical structures. They loaded the membrane permeability values and important physical parameters such as lipophilicity of these peptides onto the database.
Moreover, to make further analysis and visualization of the molecules possible, the researchers calculated the most likely 3D conformation of each peptide and added it to the database. They also encoded the chemical structure of each cyclic peptide in a novel descriptive notation (called HELM), making it possible to unambiguously refer to any cyclic peptide in the database using a short string of text.
The team has high hopes for their platform and believe that it could become a game changer in the design and development of cyclic peptide drugs. “CycPeptMPDB provides several functions, including data storage, statistics and visualization, searching and analysis, and downloading. We expect it will become a valuable tool to support membrane permeability research on cyclic peptides,” remarks Prof. Akiyama. It is also worth noting, that databases such as CycPeptMPDB are essential for training machine learning models, which can accelerate the selection of drug candidates and reveal hidden patterns in the data.
“We will continue to collect membrane permeability data of cyclic peptides and record them in CycPeptMPDB. Additionally, future improvements to the database’s online analysis platform will include an improved user-friendly interface and more integrative functions,” comments Prof. Akiyama.
Let us hope CycPeptMPDB paves the way to a future where the true potential of cyclic peptides can be exploited!
###
- Investigating the Effects on Amide-to-Ester Substitutions on Membrane Permeability of Cyclic Peptides | Tokyo Tech News
- TSUBAME supercomputer predicts cell-membrane permeability of cyclic peptides | Tokyo Tech News
- Discovering potential therapeutic protein inhibitors for Chagas disease through computational drug discovery and in vitro enzyme assays | Tokyo Tech News
- Tokyo Tech and Kawasaki City, combining forces in R&D on Computational Drug Discovery for Middle Molecules at KING SKYFRONT | Tokyo Tech News
- GHOSTZ: A faster sequence homology search algorithm based on database subsequence clustering | Tokyo Tech News
- Akiyama Laboratory
- Yutaka Akiyama | Researcher Finder – Tokyo Tech STAR Search
- Keisuke Yanagisawa | Researcher Finder – Tokyo Tech STAR Search
- Department of Computer Science, School of Computing
About Tokyo Institute of Technology
Tokyo Tech stands at the forefront of research and higher education as the leading university for science and technology in Japan. Tokyo Tech researchers excel in fields ranging from materials science to biology, computer science, and physics. Founded in 1881, Tokyo Tech hosts over 10,000 undergraduate and graduate students per year, who develop into scientific leaders and some of the most sought-after engineers in industry. Embodying the Japanese philosophy of “monotsukuri,” meaning “technical ingenuity and innovation,” the Tokyo Tech community strives to contribute to society through high-impact research.
https://www.titech.ac.jp/english/
Journal
Journal of Chemical Information and Modeling
DOI
10.1021/acs.jcim.2c01573
Method of Research
Computational simulation/modeling
Subject of Research
Not applicable
Article Title
CycPeptMPDB: A Comprehensive Database of Membrane Permeability of Cyclic Peptides
Article Publication Date
17-Mar-2023