Osaka, Japan – You can’t expect a pharmaceutical scientist to switch labs to the facilities available in a television studio and expect the same research output. Enzymes behave exactly the same. But now, in a study recently published in ACS Synthetic Biology, researchers from Osaka University have imparted an analogous level of adaptability to enzymes, a goal that has remained elusive for over 30 years.
Credit: 2022, Teppei Niide, Logistic Regression-guided Identification of Cofactor Specificity-contributing Residues in Enzyme with Sequence Datasets Partitioned by Catalytic Properties, ACS Synthetic Biology
Osaka, Japan – You can’t expect a pharmaceutical scientist to switch labs to the facilities available in a television studio and expect the same research output. Enzymes behave exactly the same. But now, in a study recently published in ACS Synthetic Biology, researchers from Osaka University have imparted an analogous level of adaptability to enzymes, a goal that has remained elusive for over 30 years.
Enzymes perform impressive functions, enabled by the unique arrangement of their constituent amino acids, but usually only within a specific cellular environment. When you change the cellular environment, the enzyme rarely functions well—if at all. Thus, a long-standing research goal has been to retain or even improve upon the function of enzymes in different environments; for example, conditions that are favorable for biofuel production. Traditionally, such work has involved extensive experimental trial-and-error that might have little assurance of achieving an optimal result.
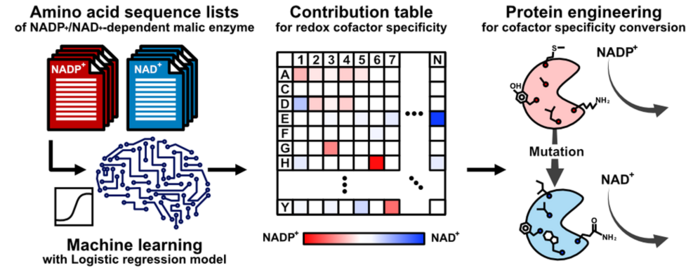
Artificial intelligence (a computer-based tool) can minimize this trial-and-error, but still relies on experimentally obtained crystal structures of enzymes—which can be unavailable or not especially useful. Thus, “the pertinent amino acids one should mutate in the enzyme might be only best-guesses,” says Teppei Niide, co-senior author. “To solve this problem, we devised a methodology of ranking amino acids that depends only on the widely available amino acid sequence of analogous enzymes from other living species.”
The researchers focused on the amino acids that are involved in the specificity of the malic enzyme to the molecule that the enzyme transforms (i.e., the substrate) and to the substance that helps the transformation proceed (i.e., the cofactor). By identifying the amino acid sequences that did not change over the course of evolution, the researchers identified the amino acid mutations that are adaptations to different cellular conditions in different species.
“By using artificial intelligence, we identified unexpected amino acid residues in malic enzyme that correspond to the enzyme’s use of different redox cofactors,” says Hiroshi Shimizu, co-senior author. “This helped us understand the substrate specificity mechanism of the enzyme and will facilitate optimal engineering of the enzyme in laboratories.”
This work succeeded in using artificial intelligence to dramatically accelerate and improve the success of substantially reconfiguring an enzyme’s specific mode of action, without fundamentally altering the enzyme’s function. Future advances in enzyme engineering will greatly benefit fields such as pharmaceutical and biofuel production that require carefully tuning the versatility of enzymes to different biochemical environments—even in the absence of corresponding enzymes’ crystal structures.
###
The article, “Logistic Regression-guided Identification of Cofactor Specificity-contributing Residues in Enzyme with Sequence Datasets Partitioned by Catalytic Properties,” will be published in ACS Synthetic Biology at DOI: https://doi.org/10.1021/acssynbio.2c00315.
About Osaka University
Osaka University was founded in 1931 as one of the seven imperial universities of Japan and is now one of Japan’s leading comprehensive universities with a broad disciplinary spectrum. This strength is coupled with a singular drive for innovation that extends throughout the scientific process, from fundamental research to the creation of applied technology with positive economic impacts. Its commitment to innovation has been recognized in Japan and around the world, being named Japan’s most innovative university in 2015 (Reuters 2015 Top 100) and one of the most innovative institutions in the world in 2017 (Innovative Universities and the Nature Index Innovation 2017). Now, Osaka University is leveraging its role as a Designated National University Corporation selected by the Ministry of Education, Culture, Sports, Science and Technology to contribute to innovation for human welfare, sustainable development of society, and social transformation.
Website: https://resou.osaka-u.ac.jp/en
Journal
ACS Synthetic Biology
DOI
10.1021/acssynbio.2c00315
Method of Research
Data/statistical analysis
Subject of Research
Not applicable
Article Title
Logistic Regression-guided Identification of Cofactor Specificity-contributing Residues in Enzyme with Sequence Datasets Partitioned by Catalytic Properties
Article Publication Date
3-Nov-2022




