Credit: Leah Stagljar
In 2011, Igor Stagljar, a professor in the University of Toronto's Donnelly Centre, came across a study that genetically linked two genes in the cell to a hard-to-treat (triple negative) breast cancer. It was not clear how the proteins encoded by these genes worked, but Stagljar had a unique way to find out.
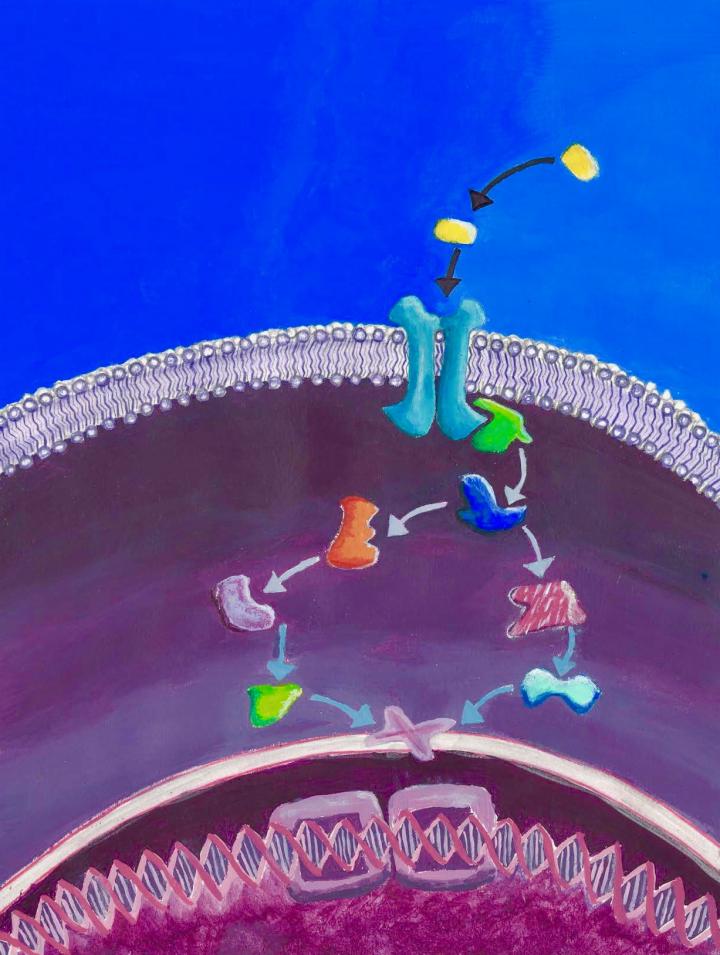
One of the proteins, called epidermal growth factor receptor (EGFR), belonged to a group called receptor tyrosine kinases (RTK), which tell the cell to grow and divide in response to signals from the cell's environment. The other one (PTPN12) was from the protein tyrosine phosphatase (PTP) class, which mainly work by shutting the RTKs down. However, the EGFR is wedged within the cell's outer envelope, or membrane, making it difficult to study by traditional methods. But with the help of MYTH and MaMTH, technologies developed in Stagljar's lab, Dr. Zhong Yao, a senior research associate in the lab, was able to show that the two proteins came into direct contact with each other. This led further support to the thinking that some breast cancers progress when the link between this particular RTK and PTP is broken, unleashing unchecked RTK signalling and, consequently, cell proliferation.
But Yao did not stop there. The high-throughput power of MYTH and MaMTH allowed him to investigate interactions between almost all human RTKs and PTPs. The resulting map charts more than RTK-PTP 300 interactions, most of which were unknown. The study will be published in the journal Molecular Cell on January 5.
"We tested interactions between almost all 58 RTKs and 144 PTPs that exist in human cells. Our map reveals new and surprising ways in which these proteins work together. These insights will help us better understand what goes wrong in cancer in order to develop more effective treatments," said Stagljar, who is also a professor in U of T's Departments of Molecular Genetics and Biochemistry.
RTKs are often mutated and hyperactive in many cancers, but only a fifth of them have been studied in enough detail to be targeted by the new generation of anti-cancer drugs called Tyrosine Kinase Inhibitors (TKIs). Yet these are some of the most promising treatments available because TKIs specifically target a mutated form of the RTK protein that's gone rogue in cancer cells, but not the normal RTK form, thus having fewer side effects than the blanket chemotherapy approach.
Lodged inside the cell membrane, with parts sticking out on either side of the cell, the RTKs act as antennae to relay signals from the outside world. When a signal–a hormone for example–arrives, an RTK attaches a phosphate group on its tail just below the cell's surface, triggering a cascade of biochemical reactions that enable the cells to grow and multiply. RTK activation is tightly controlled by the PTPs, most of which are also tethered to the membrane, whose job is to remove the phosphate group and stop the signaling cascade.
Traditional methods fall short from capturing short-lived membrane protein interactions because the surrounding membrane has to be dissolved, which changes the proteins' behaviour. Stagljar bridged this gap by developing MYTH and MaMTH which measure such fleeting interactions between membrane proteins in their natural setting in yeast and mammalian cells, respectively.
The project was done in collaboration with two leading experts in PTP biology Professors Anne-Claude Gingras, of the Lunenfeld-Tanenbaum Research institute the U of T's Department of Molecular Genetics, and Benjamin Neel, formerly of the Princess Margaret Cancer Center, UHN, currently at the New York University.
The researchers were surprised to find that some PTPs defy convention and act to promote RTK signalling suggesting that their roles are more complex than previously thought. For example, a phosphatase called PTPRA activates the EGFR, which is mutated in many cancers, revealing a new way in which cancer might spread. They also found two new phosphatases, PTPRB and PTPRH, which work as expected by grinding EGFR signalling to a halt, with potential anti-tumour properties.
"We wanted to show that these two assays, MYTH and MaMTH, that we developed in our lab, are suitable for studying these two important classes of proteins on such a large scale resulting in wealth of important data that can be used to develop new therapies against various type of cancer," said Stagljar. "Ultimately we want to build a map of interactions with all 3,000 or so human membrane proteins, of which at least 500 have direct roles in the onset of many human diseases. This will keep us busy," he added.
###
Media Contact
Jovana Drinjakovic
[email protected]
416-543-7820
@UofTNews
http://www.utoronto.ca
############
Story Source: Materials provided by Scienmag





