International team of researchers investigates how evolution forms the structure and function of a newly emerged protein in flies
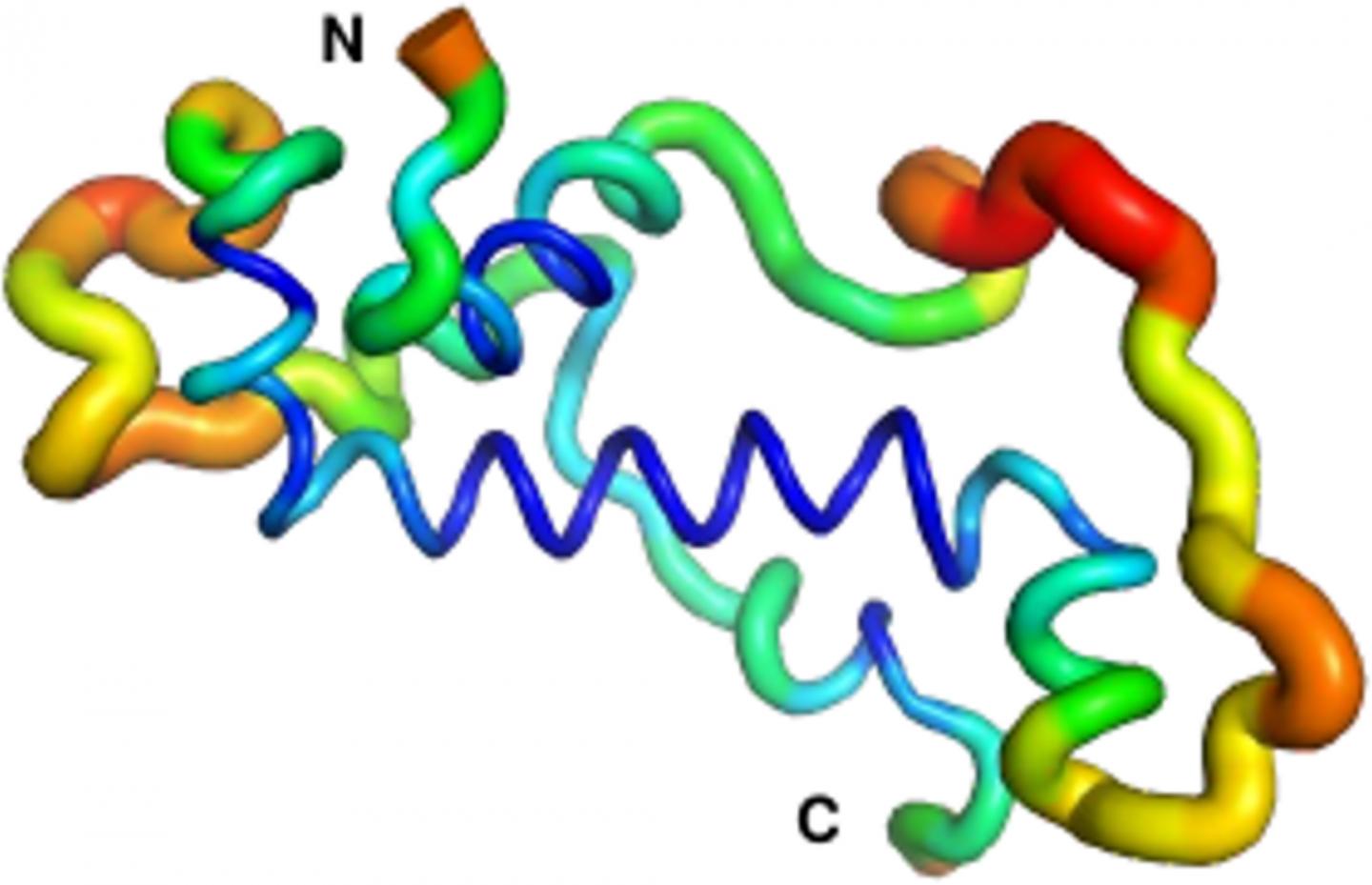
Credit: Adam Damry
Proteins are the key component in all modern forms of life. Haemoglobin, for example, transports the oxygen in our blood; photosynthesis proteins in the leaves of plants convert sunlight into energy; and fungal enzymes help us to brew beer and bake bread. Researchers have long been examining the question of how proteins mutate or come into existence in the course of millennia. That completely new proteins – and, with them, new properties – can emerge practically out of nothing, was inconceivable for decades, in line with what the Greek philosopher Parmenides said: “Nothing can emerge from nothing” (ex nihilo nihil fit). Working with colleagues from the USA and Australia, researchers from the University of Münster (Germany) have now reconstructed how evolution forms the structure and function of a newly emerged protein in flies. This protein is essential for male fertility. The results have been published in the journal “Nature Communications“.
The background:
It had been assumed up to now that new proteins emerge from already existing proteins – by a duplication of the underlying genes and by a series of small mutations in one or both gene copies. In the past ten years, however, a new understanding of protein evolution has come about: proteins can also develop from so-called non-coding DNA (deoxyribonucleic acid) – in other words, from that part of the genetic material which does not normally produce proteins – and can subsequently develop into functional cell components. This is surprising for several reasons: for many years, it had been assumed that, in order to be functional, proteins had to take on a highly developed geometrical form (a “3D structure”). It had further been assumed that such a form could not develop from a gene emerging at random, but would require a complex combination of amino-acids enabling this protein to exist in its functional form.
Despite decades of trying, researchers worldwide have not yet succeeded in constructing proteins with the desired 3D structures and functions, which means that the “code” for the formation of a functioning protein is essentially unknown. While this task remains a puzzle for scientists, nature has proven to be more adept at the formation of new proteins. A team of researchers headed by Prof. Erich Bornberg-Bauer, from the Institute of Evolution and Biodiversity at the University of Münster, discovered, by comparing the newly analysed genomes in numerous organisms, that species not only differ through duplicated protein-coding genes adapted in the course of evolution. In addition, proteins are constantly being formed de novo (“anew”) – i.e. without any related precursor protein going through a selection process.
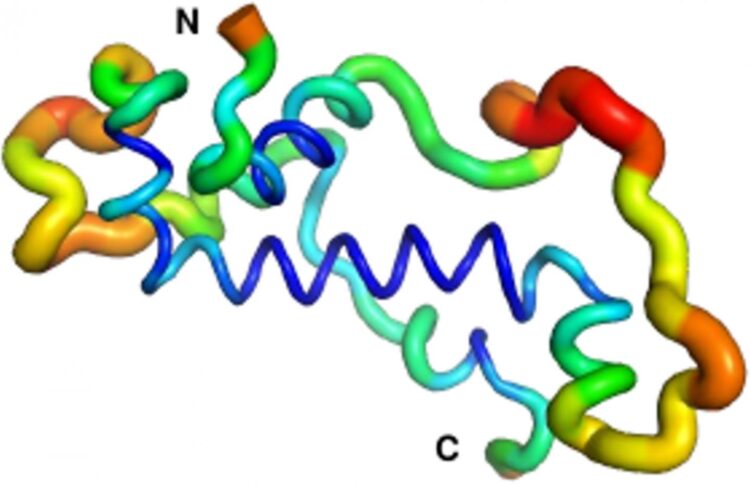
The vast majority of these de novo proteins are useless, or even slightly deleterious, as they can interfere with existing proteins in the cell. Such new proteins are quickly lost again after several generations, as organisms carrying the new gene encoding the protein have impaired survival or reproduction. However, a select few de novo proteins prove to have beneficial functions. These proteins integrate into the molecular components of cells and eventually, after millions of years of minor modifications, become indispensable. There are some important questions which many reearchers wonder about in this context: How do such novel proteins look like upon birth? How do they change, and which functions do they assume as the “new kids on the block”? Spearheaded by Prof. Bornberg-Bauer’s group in Münster, an international team of researchers has answered this question in much detail for “Goddard”, a fruit fly protein that is essential for male fertility.
Methodology
The research proceeded on three related fronts across three continents. At the College of the Holy Cross in Massachusetts, USA, Dr. Prajal Patel and Prof. Geoff Findlay used CRISPR/Cas9 genome editing to show that male flies that do not produce Goddard are sterile, but otherwise healthy. Meanwhile, Dr. Andreas Lange and PhD student Brennen Heames of Prof. Bornberg-Bauer’s group used biochemical techniques to predict the shape of the novel protein in present-day flies. They then used evolutionary methods to reconstruct the likely structure of Goddard ~50 million years ago when the protein first arose. What they found was quite a surprise: “The ancestral Goddard protein looked already very much like the ones which exist in fly species today” Erich Bornberg-Bauer explains. “Right from the beginning, Goddard contained some structural elements, so called alpha-helices, which are believed to be essential for most proteins.” To confirm these findings, the scene shifted to the Australian National University in Canberra, where Dr. Adam Damry and Prof. Colin Jackson used intensive, computational simulations to verify the predicted shape of the Goddard protein. They validated the structural analysis of Dr. Lange and showed that Goddard, in spite of its young age, is already quite stable – though not quite as stable as most fly proteins that are believed to have existed for longer, perhaps hundreds of millions of years.
The results match up with several other current studies, which have shown that the genomic elements from which protein-coding genes emerge are activated frequently – tens of thousands of times in each individual. These fragments are then “sorted” through the process of evolutionary selection. The ones which are useless or harmful – the vast majority – are quickly discarded. But those which are neutral, or are slightly beneficial, can be optimized over millions of years and changed into something useful.
###
Funding
The research undertaken received funding from the European Grant EsCat within the Horizon 2020 Research and Innovation Framework Programme No. 722610 (Münster), from the ARC Centres of Excellence in Peptide and Protein Science and Synthetic Biology (Australia), and from an NSF CAREER Grant #1652013 (USA).
Media Contact
Prof. Erich Bornberg-Bauer
[email protected]
Original Source
https:/
Related Journal Article
http://dx.