Credit: Gisou van der Goot (EPFL)
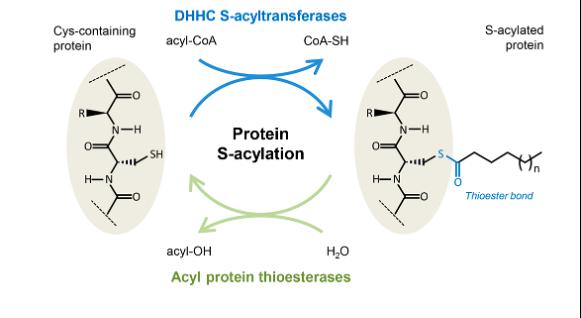
S-acylation is the process of chemically linking a lipid to protein via a thioester bond. It is an important process of the cell that regulates the localization and function of numerous proteins. It promotes lipid membrane association of the protein, for instance to the plasma membrane, Golgi apparatus, or inner nuclear membrane.
Like most biochemical processes in the cell, protein S-acylation is reversible to regulate the functions of acylated proteins. S-acylation is reversed by the enzymes acyl protein thioesterases (APTs).
To do their work, APTs have to interact with the lipid membranes that their target proteins are bound to. But even though APTs are central to the important acylation deacylation process little is known about how APTs carry out their functions.
Scientists led by Gisou van der Goot and Matteo Dal Peraro at the EPFL School of Life Sciences have now made significant inroads into our understanding of how APT2, a major acyl thioesterase in the cell, works. The work is published in Nature Chemical Biology.
First, the researchers showed that APTs have intrinsic membrane-binding capacity. Combining X-ray crystallography and molecular dynamics simulations, they showed that APTs contain in their structure positively-charged patches that allow them to electrostatically attract the lipid bilayer of a membrane.
The team also uncovered a mildly hydrophobic loop on the surface of APTs that they called the “β tongue”, which allows the enzyme to perform hydrophobic interactions with the membrane. The researchers synthesized APT2 mutants with deficient β tongues and found that they were rendered unable to bind membranes, which led them to the conclusion that the ability of APT2 (and other thioesterases by extension) to bind membranes is mediated by the ß tongue sequence.
Analyzing the structure of APT2, the researchers also identified a site that can lead to its degradation. The site allows the enzyme to be bound by ubiquitin, a protein that the cell uses to mark molecules for breakdown. Essentially, APT2 contains a built-in control mechanism for its own degradation – but this can only happen after its target has been deacylated and APT2 has been released from it. Alternatively, after one job is complete, APT2 can relocate to another membrane, bind it, and deacylate another protein there.
The researchers then turned to the S-acylation of APT2 itself. Previous studies have shown that the enzyme is heavily accumulated in the cell’s Golgi apparatus, which is the organelle that packages new protein into vesicles before sending them off to the cell’s membrane.
Using another APT2 mutant, the researchers were able to determine that this accumulation depends on the S-acylation of APT2 itself – on a cysteine amino acid along its sequence (Cys2). In short, S-acylation on Cys-2 is essential for APT2 to be able to stably bind lipid membranes and deacylate its targets in the cell.
The team then searched for possible candidate enzymes that can acylate APT2. To do this, they screened all palmitoyltransferase enzymes. The results showed that APT2 can be S-acylated by either of two palmitoyltransferases, ZDHHC3 or ZDHHC7.
Finally, the scientists brought their data together to work out how APT2 actually binds lipid membranes, which is essential to its capacity to perform its function in the cell.
What they found was that APT2 binds membranes in a three-step process. First, long-range electrostatic interactions attract the enzyme, through its positive patches, to the lipid membrane. There, the β tongue “dips” into the membrane and holds APT2 temporarily in place, which is necessary for it to be “met” by the enzymes that will acylate it. This leads to APT2 being stably bound to the membrane and ready to perform its deacetylating functions.
“This study shows that APT2 is in fact a hybride between a lipid carrier protein, which can extract lipid from membranes, and a hydrolase, which can cut the lipid of a protein,” says Gisou van der Goot.
###
Other contributors
EPFL Protein Production and Structure Core Facility
Swiss Institute of Bioinformatics
Reference
Laurence Abrami, Martina Audagnotto, Sylvia Ho, Maria Jose Marcaida, Francisco S. Mesquita, Muhammad U. Anwar, Patrick A. Sandoz, Giulia Fonti, Florence Pojer, Matteo Dal Peraro, F. Gisou van der Goot. Molecular mode of action of an Acyl Protein thioesterase. Nature Chemical Biology 11 March 2021. DOI: 10.1038/s41589-021-00753-2
Media Contact
Nik Papageorgiou
[email protected]
Related Journal Article
http://dx.




