Rice’s programmable CRISPR/Cas9-based kinase offers insights into, control over regulatory histone proteins
Credit: Hilton Lab/Rice University
HOUSTON – (Feb. 9, 2021) – Finding a needle in a haystack is hard enough. But try finding a specific molecule on the needle.
Rice University researchers have achieved something of the sort with a new genome editing tool that targets the supporting players in a cell’s nucleus that package DNA and aid gene expression. Their work opens the door to new therapies for cancer and other diseases.
Rice bioengineer Isaac Hilton, postdoctoral researcher and lead author Jing Li and their colleagues programmed a modified CRISPR/Cas9 complex to target specific histones, ubiquitous epigenetic proteins that keep DNA in order, with pinpoint accuracy.
The open-access research appears in Nature Communications.
Histones help regulate many cellular processes. There are four in each nucleosome (the basic “beads on a string” in DNA) that help control the structure and function of our genomes by exposing genes for activation.
“Nucleosomes serve as architectural substrates to fit our DNA inside of our cells, and can also control access to key parts of our genomes,” Hilton said.
Like other proteins, histones can be triggered by phosphorylation, the addition of a phosphoryl group that can control protein-protein or protein-DNA interactions.
“Histones can display an exquisitely diverse spectrum of chemical modifications that serve as beacons or regulatory markers and tell which genes to turn on, and when, and how much to do so,” Hilton said. “One of these mysterious modifications is phosphorylation, and we aimed to better illuminate the mechanism by which it can rapidly turn human genes on and off.”
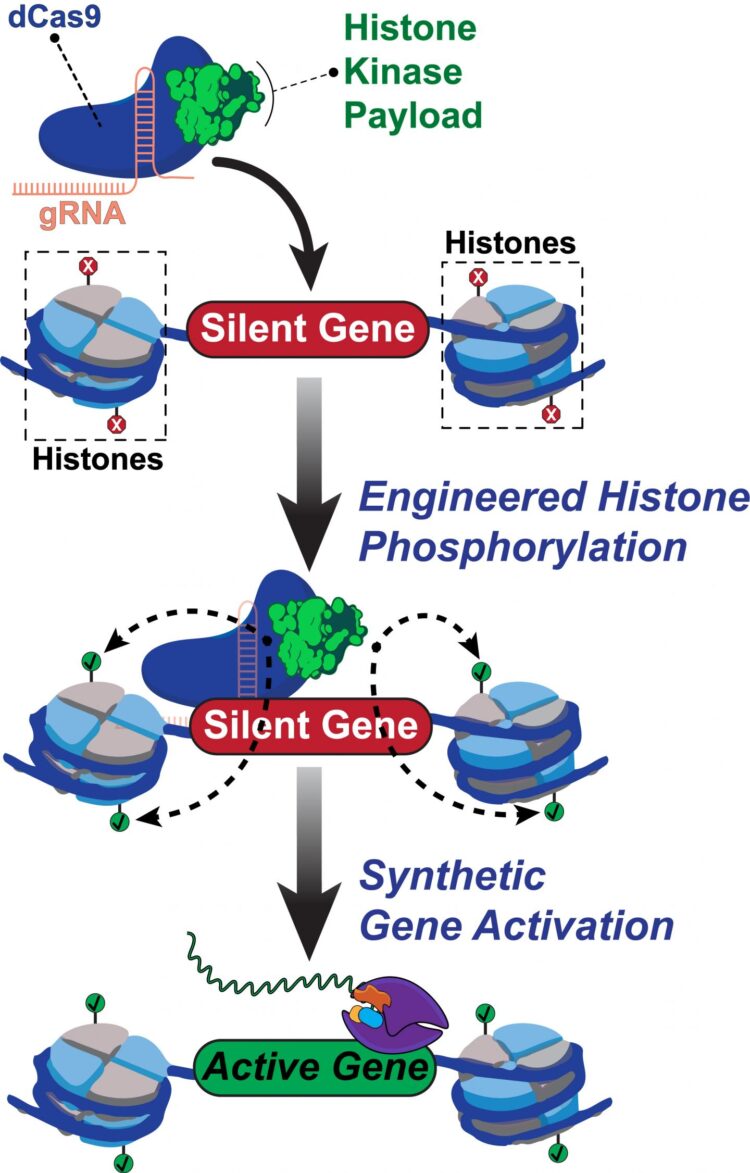
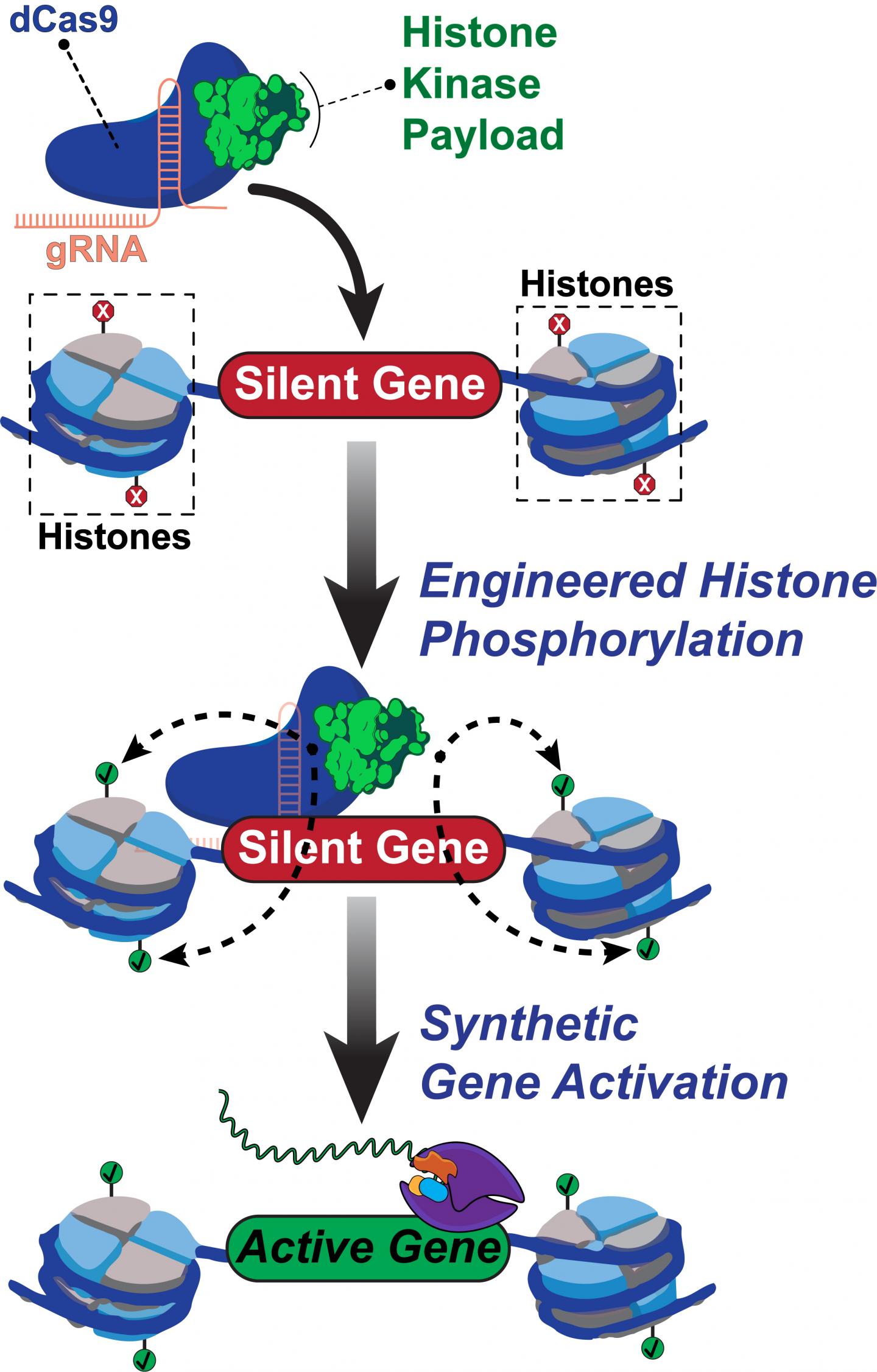
No other epigenome editing technique has enabled site-specific control over histone phosphorylation, he said. The programmable Rice tool, called dCas9-dMSK1, fuses a deactivated “dCas9” protein and a “hyperactive” human histone kinase, an enzyme that catalyzes phosphorylation.
CRISPR/Cas9 typically employs guide RNAs and Cas9 “scissors” to target and cut sequences in DNA. The new tool programs deactivated dCas9 to target without cutting sequences, instead using the recruited dMSK1 enzyme to phosphorylate the targeted histone and turn on nearby genes.
The researchers used dCas9-dMSK1 to uncover novel genes and pathways that are pivotal for drug resistance. Li used it to identify three genes previously linked to melanoma drug resistance. “And then she identified seven new genes linked to melanoma resistance,” Hilton said. “It’s an exciting finding that we are following up on.
“Histone proteins that wrap up DNA can have all sorts of chemical marks and combinations on them,” he said. “This results in what has been dubbed a histone code, and one of our goals is to work to decipher it.”
Li’s tool also confirms how specific histone marks communicate with one another. “It tells us that chemical modifications on histones talk to each other, and we can show it happening at specific spots in the human genome,” Li said. “And that’s linked to a gene turning on, so this allows us to synthetically control them.”
Li said a long-term goal is to target a range of other histone marks. “It’s a complicated story,” she said. “There are a lot of different positions and features of histones that we want to study.”
“Getting these technologies into patients is a long process,” Hilton added. “But tools like this are the first step and can pave the way towards understanding how normal cellular processes unfortunately go awry in human diseases.”
###
Co-authors of the paper are Rice postdoctoral researcher Barun Mahata, graduate students Mario Escobar, Jacob Goell and Kaiyuan Wang and undergraduate Pranav Khemka. Hilton is an assistant professor of bioengineering and biosciences.
The Cancer Prevention and Research Institute of Texas supported the research.
-30-
Read the paper at https:/
This news release can be found online at https:/
Follow Rice News and Media Relations via Twitter @RiceUNews.
Additional Contact:
Mike Williams
713-348-6728
[email protected]
Related materials:
CPRIT grant brings epigenetics researcher to Rice:
http://news.
Rice lab gives structure to histone discovery:
http://news.
Chromosome organization emerges from 1D patterns:
http://news.
Hilton Lab:
https:/
Rice Department of Bioengineering:
https:/
Rice Department of BioSciences:
https:/
Images for download:
https:/
Rice University scientists built a new tool to engineer and understand how human genes are turned on. The team created a synthetic two-part protein based on dCas9 and a modified enzyme called dMSK1 to deliver chemical payloads at precise spots near human genes. The tool causes pinpoint changes to histone marks and with the help of other proteins, the activation of silent human genes.
(Credit: Hilton Lab/Rice University)
https:/
CAPTION: Isaac Hilton.
(Credit: Rice University)
https:/
CAPTION: Jing Li.
(Credit: Rice University)
Located on a 300-acre forested campus in Houston, Rice University is consistently ranked among the nation’s top 20 universities by U.S. News & World Report. Rice has highly respected schools of Architecture, Business, Continuing Studies, Engineering, Humanities, Music, Natural Sciences and Social Sciences and is home to the Baker Institute for Public Policy. With 3,978 undergraduates and 3,192 graduate students, Rice’s undergraduate student-to-faculty ratio is just under 6-to-1. Its residential college system builds close-knit communities and lifelong friendships, just one reason why Rice is ranked No. 1 for lots of race/class interaction and No. 1 for quality of life by the Princeton Review. Rice is also rated as a best value among private universities by Kiplinger’s Personal Finance.
Media Contact
Jeff Falk
[email protected]
Original Source
https:/
Related Journal Article
http://dx.