microRNAs (miRNA) are recently discovered, short segments of RNA, which play vital roles in health and disease. microRNA over- or under-abundance can switch critical disease genes on or off, particularly…
Credit: The Biodesign Institute at Arizona State University
As the floor plan of the living world, DNA guides the composition of animals ranging from unicellular organisms to humans. DNA not only helps shepherd every organism from birth through death, it also plays an essential role in the development of many human diseases.
But it wasn't always so. Long before DNA emerged as the molecule of life, its closely related cousin, RNA (ribonucleic acid), held center stage.
The RNA world refers to a time in earth's distant past when primitive forms used RNA rather than DNA to archive genetic information, pass it along using RNA-based copying machinery and perform biological reactions.
With the emergence of DNA, RNA came to play an intermediary role, copying DNA messages known as genes and translating them into proteins. This pathway from DNA to RNA to protein has become so engrained in the field of biology it is often referred to as "the central dogma."
Recently, however, RNA's strict subservience to DNA has been called into question. New discoveries have prompted an explosion in RNA research, with vital implications for both the foundations of biology and the practice of medicine. (Sidney Altman, who won the Nobel Prize for establishing that RNA can act independently and perform chemical reactions on its own, providing powerful evidence for the RNA world hypothesis, has recently joined ASU's School of Life Sciences).
RNA resurgence
In the last few years, small snippets of RNA, which may have played a key role in the planet's earliest flickering of life, have been uncovered and examined in great detail. Their discovery, first in the tiny soil-dwelling nematode worm C. elegans and shortly thereafter, across the web of life, marks a revolution in biology, with broad implications in the fight against nearly every known disease.
These abbreviated RNA fragments, known as microRNA (miRNA) are composed of just 18-22 nucleotides. They are too short to code for proteins, (in the manner of longer messenger RNA or mRNA strands). Instead, they act as a subtle and extremely sophisticated network of gene regulators.
To accomplish this, miRNAs find complementary sections of mRNA targets and bind with a specific location known as the 3'UTR (for UnTranslated Region). After binding to the UTR, the miRNA inactivates mRNA, interfering with its translation to protein.
In a new study, Marco Mangone, a researcher at the Biodesign Center for Personalized Diagnostics and his colleagues conduct the largest screening of miRNAs to date. Their findings, which appear in the advanced online issue of the journal Genome Research, point to a new understanding of these enigmatic molecules and their vital participation in organismic development, the maintenance of health and the development of disease.
"MicroRNA are very mysterious. They are really relics of the RNA world–pieces of RNA that are highly reactive, very small and which pair and bind other RNA and facilitate catalytic reactions," Mangone says. "We don't know much about them–where they come from or how they regulate gene expression, but they are very misregulated in many diseases."
Where it started
The miRNA story first gathered momentum in 1993, when a particular RNA fragment in C. elegans, known as lin-4, was characterized molecularly. (C. elegans is a model organism whose genome has been fully sequenced. The worm's simple body plan, well-studied genetics and characteristic developmental stages have provided researchers with a wealth of information, much of it applicable to higher organisms, including humans.)
Intriguingly, the gene yielding lin-4 doesn't code for a protein, but rather, for this tiny piece of RNA–the first miRNA discovered. Originally, lin-4 was viewed as a curiosity specific to C. elegans, but soon, a second miRNA, let-7, was discovered, and this one was found to be evolutionarily conserved, meaning that it recurs across the web of life forms, including, humans.
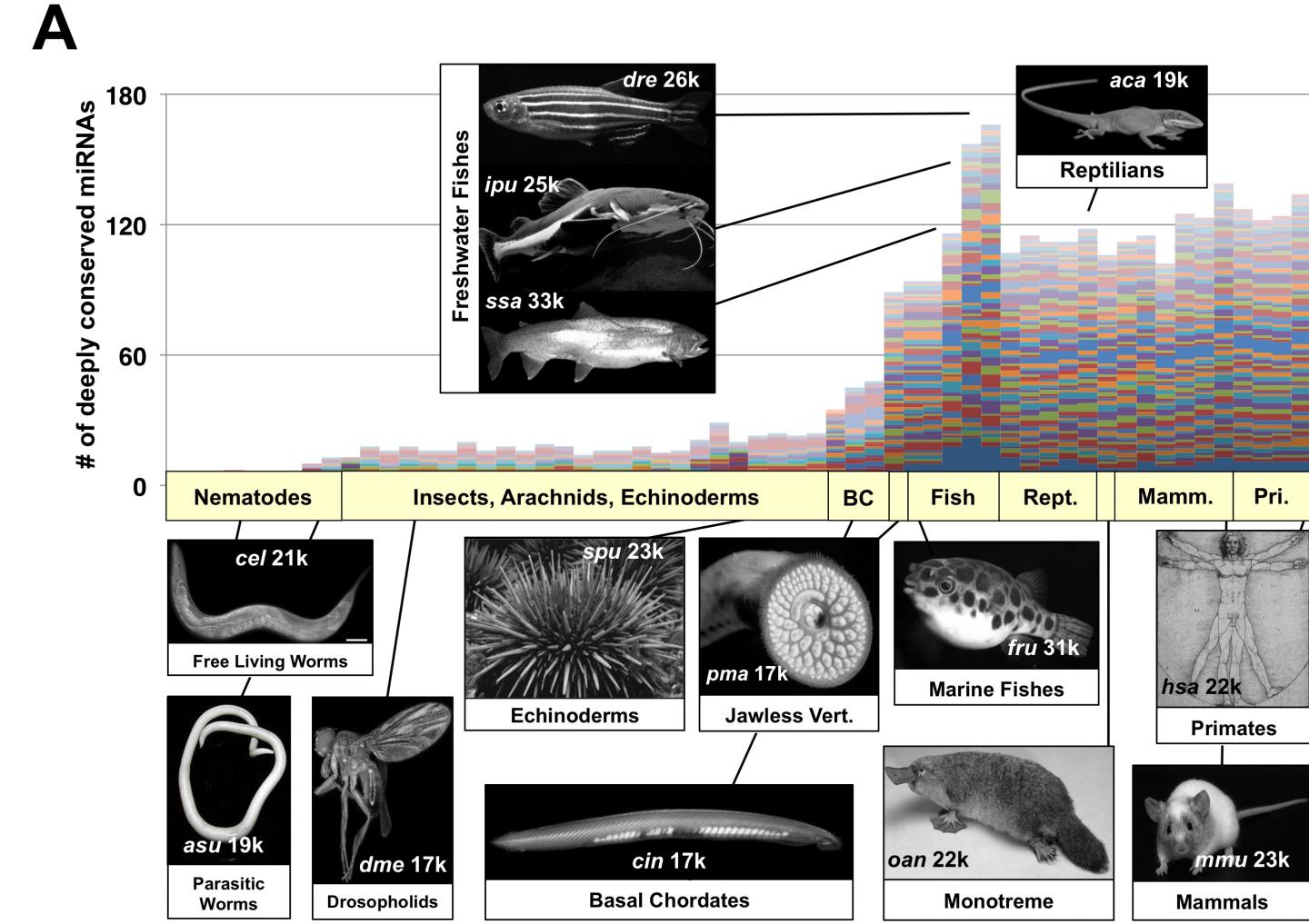
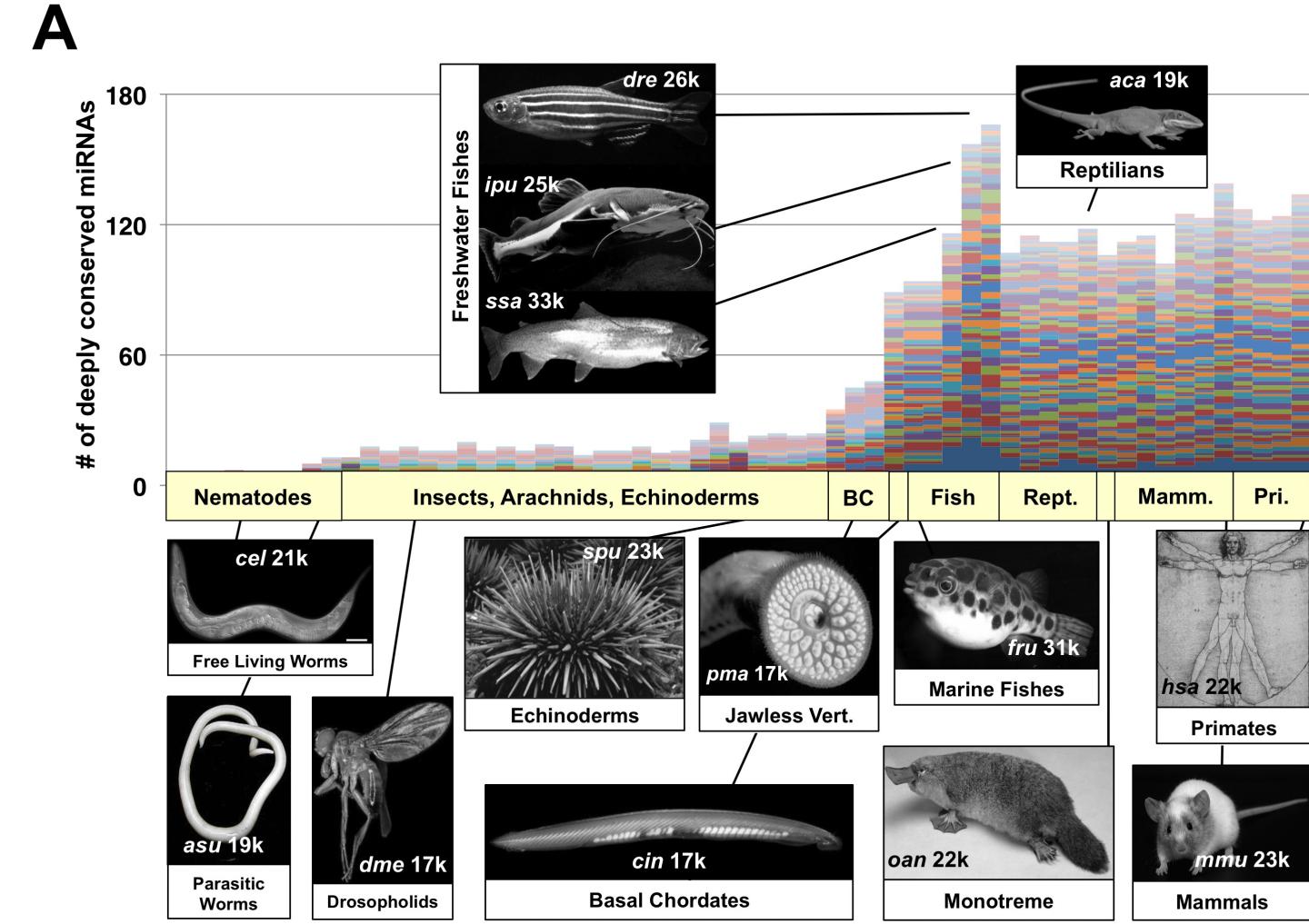
Over the course of evolution, higher species have become heavily invaded by miRNA, which have undergone radical expansion in vertebrates, while the number of genes has remained stable. Astonishingly, worms, flies, and humans all have roughly 20,000 protein coding genes, though humans have far more miRNA than simpler species. Mangone insists this intriguing puzzle begs for a convincing explanation.
"What we found is that these miRNAs started parasitizing cells because they needed to target and regulate more gene expression, modulating entire pathways not just single genes," Mangone says.
Unseen universe
Various complex networks act between transcription (from DNA to RNA) and translation (from RNA to protein). Until recently, however, this biological dark matter has eluded study. "So far, we haven't known the contribution of post-transcriptional gene regulation in any disease. We don't understand the rules about how these miRNAs regulate these targets," Mangone says. "With this paper, we can now begin to understand the sophistication involved in the miRNAs control of gene expression."
These tiny strips of RNA, many with very similar nucleotide sequences, form miRNA families, capable of targeting disparate gene pathways. While these miRNA families were originally thought to provide redundancy and robustness in gene regulation, the new study suggests a different explanation for miRNA abundance and proliferation in higher species.
The research findings show that miRNA not only target specific genes but expand their network of action through minor mutations in specific portions of their sequence, which Mangone named "evolutionary hotspots." This allows particular miRNA family members to extend their range of modulating activity while maintaining a common set of core targets.
Regulation of gene pathways has profound implications for human disease. As one example, a miRNA known as miR-10b is highly abundant in early stage breast cancer, particularly during the phase when diseased cells enter the bloodstream. Mangone's group discovered that this miRNA targets many genes involved in the retinoic acid pathway, which plays a crucial role in development, overseeing cell differentiation, proliferation, and cell death. Dysregulation of retinoic signaling can contribute to serious disease.
miR-10b is now recognized as an oncogene–over-expressed in breast cancer. The effect of this over-expression is to silence specific genes that can force the cell to induce senescence and cell cycle arrest. With these critical genes silenced by miRNA, cells continue endless rounds of cycling–a hallmark of cancer. As Mangone explains, "If you could remove miR-10b you could eventually reactivate these cells and then when you add retinoic acid or hormones you can actually force the cell to induce cell death, senescence, and arrest. This is the next step."
Target rich environment
Ferreting out miRNAs and their targets has been the focus of Mangone's research. In earlier work, he created a library of 20,000 3'UTRs in C. elegans–potential mRNA targets for the miRNAs. At ASU he developed the first publically available human 3'UTR clone library, which represents about 10 percent of the human genome.
Mangone also developed at ASU an innovative, high-throughput approach to detect miRNA targets in genes. Mangone's called this technology 3'LIFE assay (Luminesce-based Identification of Functional Elements in 3'UTRs), and his group was able to screen serveral disease-relevant miRNAs against the large UTR library, using florescent labeling to identify each miRNA binding event. "This high-throughput screening for functional targets reveals that each miRNA regulates a large number of genes with cooperative function in a regulatory network," Mangone says.
The results of the study are thought-provoking and run counter to previously proposed ideas about the nature and function of miRNAs. The study explores miRNA families, finding that the mutations that give rise to them do not occur randomly, with respect to either sequence location or function. Rather, mutations occur in preferential regions–evolutionary hotspots–indicating the mutations are functional and maintained through evolution. Contrary to the common conception of single miRNAs regulating single genes, the study observed that each miRNA tends to control multiple members in the same regulatory pathways.
Finally, the study explores the specific sequence locations in miRNAs known as the seed regions, often assumed to be essential for binding with the 3'UTRs. "We show in this manuscript that there are a lot of targets that do not have the seed region, yet they are still targeted, suggesting that just the presence of the seed region in the 3' UTR doesn't correlate with the binding on the miRNA. There are other rules, but now we begin to understand the language that these miRNAs speak."
Mangone hopes to eventually screen the entire human genome to complete the map of miRNA activity. Ongoing miRNA research will continue to address central questions in biology and may have profound implications for drug discovery.
###
Media Contact
Richard Harth
[email protected]
504-427-2666
@ASU
http://asunews.asu.edu/
############
Story Source: Materials provided by Scienmag