Credit: Nature Communications, Tokyo Tech
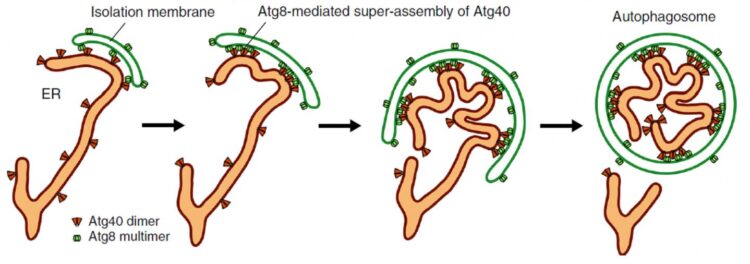
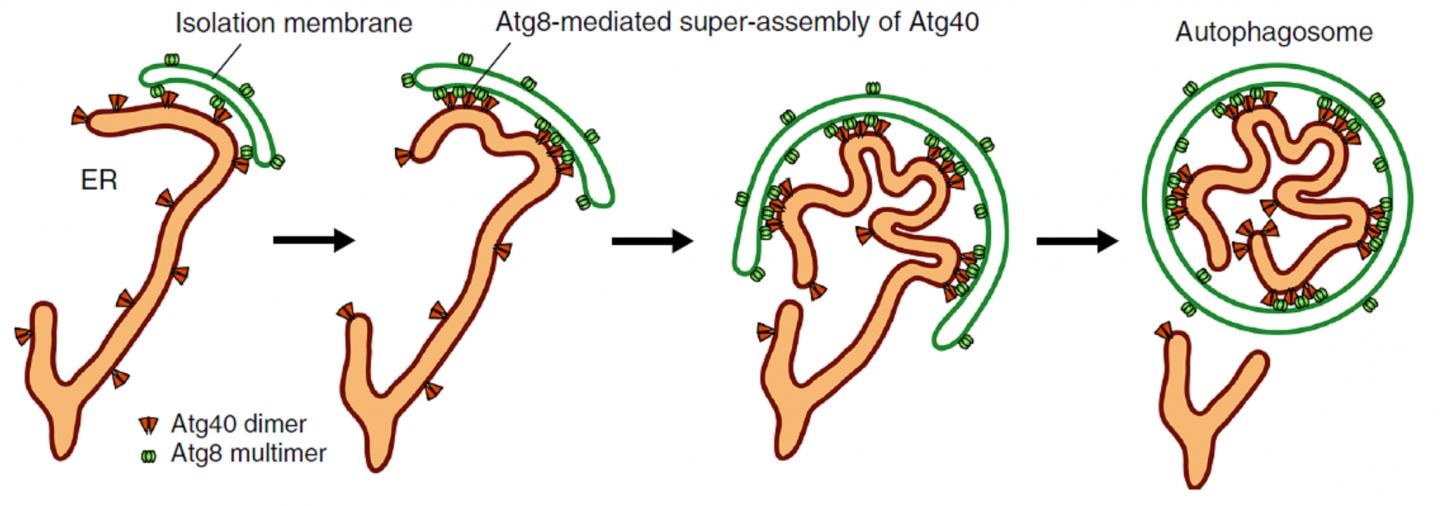
The endoplasmic reticulum (ER) is an important part of eukaryotic cells (the type of cells that make up every living thing other than bacteria or viruses, including humans). They are a mass of tubes connected to the nucleus of the cell; the production of both proteins and lipids occur in the networks of the ER. For this organelle to properly function, cells routinely degrade portions of the ER so that it can be renewed. This process is called ER autophagy, or ER-phagy, where a structure called an “isolation membrane” expands and closes up to form an “autophagosome.” The closure isolates various cellular materials including the ER within the autophagosome, which then transports the waste away for degradation.
While this process can be random, scientists have uncovered “autophagy receptors” that bind specifically to certain targets and interact with a group of proteins called Atg8, located on the isolation membrane. This interaction allows cells to target specific parts for degradation. In yeast, an organism commonly used for biological research, scientists have identified the protein Atg40 as an ER-phagy receptor, and also found that parts of its structure share similarities to a group of proteins called DP1/Yop1 (reticulon-like proteins), which “curves” the ER membranes into shape and maintains their tubular structures.
“Our previous work reveals that Atg40 is important for ER-phagy, but we actually know very little about how the process works,” explained Dr. Hitoshi Nakatogawa of the Tokyo Tech, who led a team of scientists in research that investigated the mechanisms of Atg40 involvement in ER-phagy. “Because degradation of ER is so important for proper cellular function, gaining a better understanding of ER-phagy will improve basic biological knowledge.”
Their experiments with yeast, the findings of which are published in Nature Communications, showed that Atg40 is important for “curving and folding” the ER membrane, and therefore has a similar function to DP1/Yop1, explaining their structural similarities. Atg40 is also necessary for ER-phagy, specifically being involved in breaking up the ER membrane so that it can be imbibed by autophagosomes. Researchers demonstrated that during this folding and fragmentation of the ER, Atg40 forms a protein assembly (cluster of proteins) by interacting with Atg8 located specifically at points of contact between the ER and the isolation membrane (as shown in Figure 1). In other words, Atg40 does not randomly or always remodel ER structure; it does so only for the ER parts that will be degraded.
Regarding the significance of these results, Dr. Nakatogawa commented: “What I find particular exciting is the insight we gained on a crucial part of how cells work, how they deal with waste or get rid of abnormal cell parts. Our work doesn’t just have implications for ER-phagy though, it can also potentially tell us something about how other organelles, like the nucleus or mitochondria, are degraded.”
Besides just being valuable basic research, these findings also have significant practical applications. Knowing the mechanisms of organelle degradation might help the development of drugs that target this process if it breaks down. This presents potential attractive solutions for diseases involving the malfunction of ER such as sensory neuropathy.
###
Media Contact
Kazuhide Hasegawa
[email protected]
Original Source
https:/
Related Journal Article
http://dx.