Credit: Y.-X. Ren, J. Wu, Q. T. K. Lai, H. M. Lai, D. M. D. Siu, W. Wu, K. K. Y. Wong, and K. K. Tsia
An arsenal of advanced microscopy tools is now available to provide high-quality visualization of cells and organisms in 3D and has thus substantiated our understanding the complex biological systems and functions.
In a new paper published in Light: Science & Applications, a research team led by the University of Hong Kong (HKU) developed a new form of imaging modality, coined coded light-sheet array microscopy (CLAM) that allows full 3D parallelized fluorescence imaging without any scanning mechanism – a capability that is otherwise challenging in the existing techniques.
Established 3D biological microscopy techniques, notably confocal, multiphoton microscopy, and light-sheet fluorescence microscopy (LSFM), predominantly rely on laser-scanning for image capture. Yet, it comes at the expense of imaging speed because the entire volume has to be sequentially scanned point-by-point, line-by-line or plane-by-plane at a speed limited by the mechanical motions involving the imaging parts.
Even worse, many serial scanning approaches repeatedly excite out-of-focus fluorescence, and thus accelerate photobleaching and photodamage. They are thus not favorable for long-term, large-scale volumetric imaging critically required in applications as diverse as anatomical science, developmental biology and neuroscience.
3D parallelization in CLAM requires even gentler illumination to achieve a similar level of image sensitivity at the same volumetric frame rate. Hence, it further reduces the photobleaching rate and thus the risk of photodamage. This is a critical attribute for preserving the biological specimen viability in long term monitoring studies.
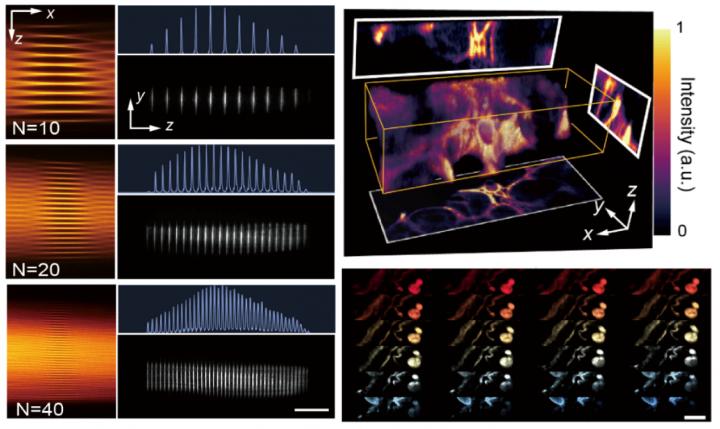
The heart of CLAM is the concept of ‘infinity mirror” (i.e., a pair of parallel mirrors), which is common in visual art and decoration, and has previously been adopted by the same team for enabling ultrafast optofluidic single-cell imaging. Here the team employed the ‘infinity mirror” together with simple beam shaping to transform a single laser beam into a high-density array of few tens of light-sheets for 3D parallelized fluorescence excitation.
“One distinct feature of CLAM is its ability to flexibly reconfigure the spatial density and temporal coherence of the light sheet array, simply by tuning the mirror geometry, such as mirror separation and tilt angle.” explained Dr. Yuxuan Ren, the postdoctoral researcher and the first author of the work.
“This capability has been challenging in the existing coherent wavefront shaping methods, yet could allow efficient parallelized 3D LSFM in scattered tissue imaging with minimal speckle artifact.” Ren added.
CLAM also adopts code division multiplexing (CDM) (e.g., orthogonal frequency division multiplexing demonstrated in this work), a technique widely used in telecommunication, to imprint the fluorescence signal from each image plane with a unique code. As a result, it allows parallelized 3D image capture with optical sectioning by using a 2D image sensor.
“CLAM has no fundamental limitation in scaling to higher volume rate as camera technology continually advances,” Dr. Kevin Tsia, Associate Professor in Department of Electrical and Electronic Engineering at HKU and the leading researcher of the team pointed out.
“Also, CLAM can be adapted to any existing LSFM systems with minimal hardware or software modification. Therefore, it is readily available for dissemination to the wider community of LSFM and related 3D imaging techniques.” added Tsia.
###
The team is planning to further upgrade the current CLAM system for applications involving long-term dynamical volumetric imaging of live cellular, tissue, and organism, as well as high-throughput volumetric visualization for 3D histopathological investigation of archival biological samples.
This research received funding from the Research Grants Council of the Hong Kong Special Administrative Region of China, Innovation and Technology Support Program, the University Development Funds of the University of Hong Kong, and Natural Science Foundation of China.
Media Contact
Yuxuan Ren
[email protected]
Related Journal Article
http://dx.




