Credit: Max Planck Florida Institute for Neuroscience
The power and convenience of modern-day word-processing programs, like Microsoft Word, have revolutionized our daily tasks. Need to create a quick resume for a new job opportunity? Procrastinating on that final term paper due tomorrow? Even creating a quick grocery list- Most of us rely on word-processing programs as stewards of our written lives. The functionality is impressive and unlike its archaic predecessor, the type writer, just a few keystrokes can change, delete, or add words as the user desires.
For years researchers have been searching for ways to mimic these capabilities but on the genetic scale, in order to accurately and efficiently alter genetic information. Developing a “Microsoft Word of DNA”, would allow scientists the opportunity to better study the functioning of individual genes and the mutations that contribute to genetic disorders. With a single discovery, called CRISPR-Cas9, scientists were forever upgraded from DNA typewriters to a DNA word-processor; editing genes with the same ease, precision and versatility. The technology has gained immense traction and popularity due to its flexibility and customizable nature.
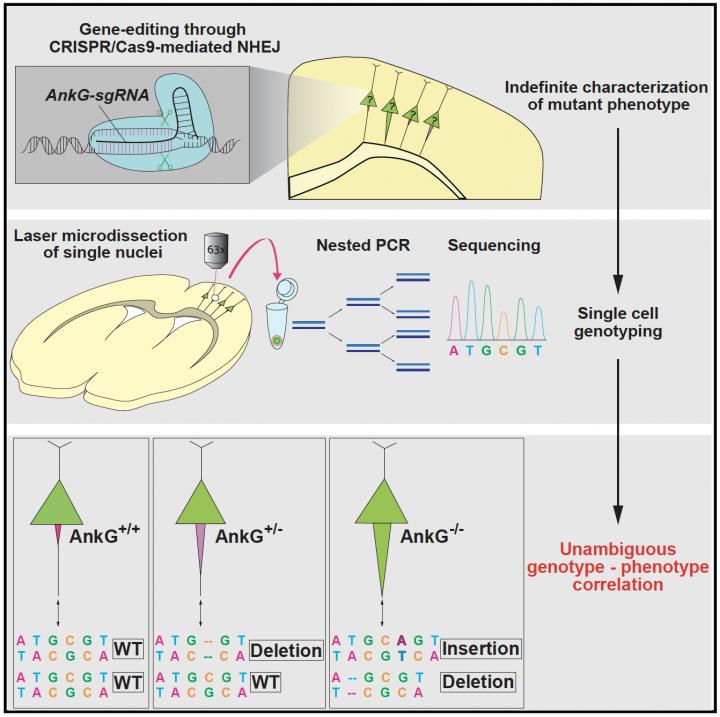
Though powerful, CRISPR is far from a perfect technique and like any other, has its drawbacks and limitations. One of its primary uses, involves precisely targeting and mutating a gene of interest so that it no longer functions within a specific type of cell. When a gene is rendered nonfunctional, any characteristic changes to the cell (known as phenotypes) can be studied in order to get a better picture of what that particular gene does. But the way CRISPR mutates individual genes can pose a challenge to researchers. When placed in a cell, the CRISPR-Cas9 system precisely mutates a targeted gene by cutting the cell’s DNA. The cell then repairs its broken DNA predominantly through a process called nonhomologous end-joining (NHEJ). But this repair process is error prone and can cause variability in the repaired DNA; often leading to substitutions, deletions or additions to the genetic code. Adding to the challenge is that the efficiency of CRISPR can vary, acting to render both copies of a targeted gene nonfunctional or sometimes only one. These unknown CRISPR caused DNA changes can make it extremely difficult for scientists to interpret the underlying genetic cause of an observed phenotype; making the tool far less useful.
In a recent publication in Cell Reports, the Taniguchi Lab at the Max Planck Florida Institute for Neuroscience (MPFI) have developed a new methodology that allows the linking of phenotype to genotype. Innovatively combining the cutting-edge technique of laser microdissection with single cell genotyping, the Taniguchi Lab has designed an experimental pipeline capable of studying CRISPR mediated effects in cells while accurately ascertaining the exact DNA changes that caused them. This novel protocol will open up new avenues of study for neurobiology and further upgrade the already powerful abilities of CRISPR.
“Though CRISPR precisely targets a gene of interest, due to NHEJ, its effects can be highly variable,” explains Andre Steinecke, Ph.D., Research Fellow and first author of the publication. “CRISPR can leave cells with either fully nonfunctional genes, weakened genes or sometimes even enhance their function. This isn’t such a problem when removing one that causes a very noticeable effect in cells because you can easily visualize the change and absence of the protein coded by the gene. But some, especially genes in the brain, don’t have strikingly obvious effects or are very difficult to visualize. Our goal was to create a widely applicable strategy, capable of reliably determining the exact genetic cause and correlate it to observed phenotype.”
To validate their strategy, the team at MPFI designed CRISPR technology to target a gene in pyramidal neurons encoding a critical structural protein, called Ankyrin-G (AnkG). Normally, the AnkG protein is confined to a specialized region of the neuron known as the axon initial segment (AIS), which is responsible for generating action potentials. When AnkG is removed, the AIS undergoes a noticeable thickening that can be detected using microscopy. With this characteristic feature, neurons that lack AnkG could be readily distinguished and their exact genotype could then be confirmed. They found that predominately, neurons transfected with their CRISPR probe exhibited a loss of AnkG as well as substantially thickened AIS. But a small portion of neurons transfected with CRISPR still exhibited AnkG levels and AIS thickness comparable with wildtype neurons; demonstrating the varying effects of CRISPR on different cells. To probe and confirm the underlying genetic causes, the team then used laser microdissection to isolate and extract individual neurons whose phenotype had already been characterized. Once extracted, the team sequenced each individual cell separately to determine the genotype. They found that their strategy could reliably and reproducibly link observed phenotype to genotype, where neurons lacking AnkG with thickened axons showed loss-of-function mutations in both copies of the gene whereas neurons with normal levels of AnkG either showed mutations in only one copy (neurons transfected with CRISPR) or normal genotypes (control neurons). The team then confirmed their strategy using two additional genes, MeCP2 and Satb2, finding that their process could once again effectively correlate observed feature to underlying genetics.
“CRISPR/Cas9-based gene targeting holds great promise for systematic understanding of the molecular basis underlying the assembly, function, and dysfunction of neural circuits,” notes Hiroki Taniguchi, Ph.D. “The perfect matching between genotypes determined by our single cell sequencing and those deduced from phenotype evaluation, suggests that our approach is a powerful new method capable of enhancing the reliability and expanding the applications of CRISPR-based techniques.”
###
Media Contact
Helena Decker
[email protected]
Related Journal Article
http://dx.




