Credit: Jijie Chai
Although separated by more than one billion years of evolution, plants and animals have hit upon similar immune strategies to protect themselves against pathogens. One important mechanism is defined by cytoplasmic receptors called NLRs that, in plants, recognize so-called effectors, molecules that invading microorganisms secrete into the plant’s cells. These recognition events can either involve direct recognition of effectors by NLRs or indirect recognition, in which the NLRs act as ‘guards’ that monitor additional host proteins or ‘guardees’ that are modified by effectors. Host recognition of effectors, whether direct or indirect, results in cell death to confine microbes to the site of infection. However, until now, a detailed understanding of the mechanisms of action of plant NLRs has been lacking, and much of our understanding of how these molecules function in plants has been based on comparison with animal counterparts.
In two new studies published in the journal Science, Jijie Chai who is affiliated with Tsinghua University in Beijing as well as the University of Cologne and the Max Planck Institute for Plant Breeding Research together with the groups of Hong-Wei Zhang and Jian-Min Zhou at Tsinghua University and the Chinese Academy of Sciences in Beijing have now pieced together the sequence of molecular events that convert inactive NLR molecules into active complexes that provide disease resistance.
The authors focused their attentions on a protein called ZAR1, an ancient plant molecule that is likely to be of broad importance since it interacts with multiple ‘guardees’ to recognize unrelated bacterial effectors.
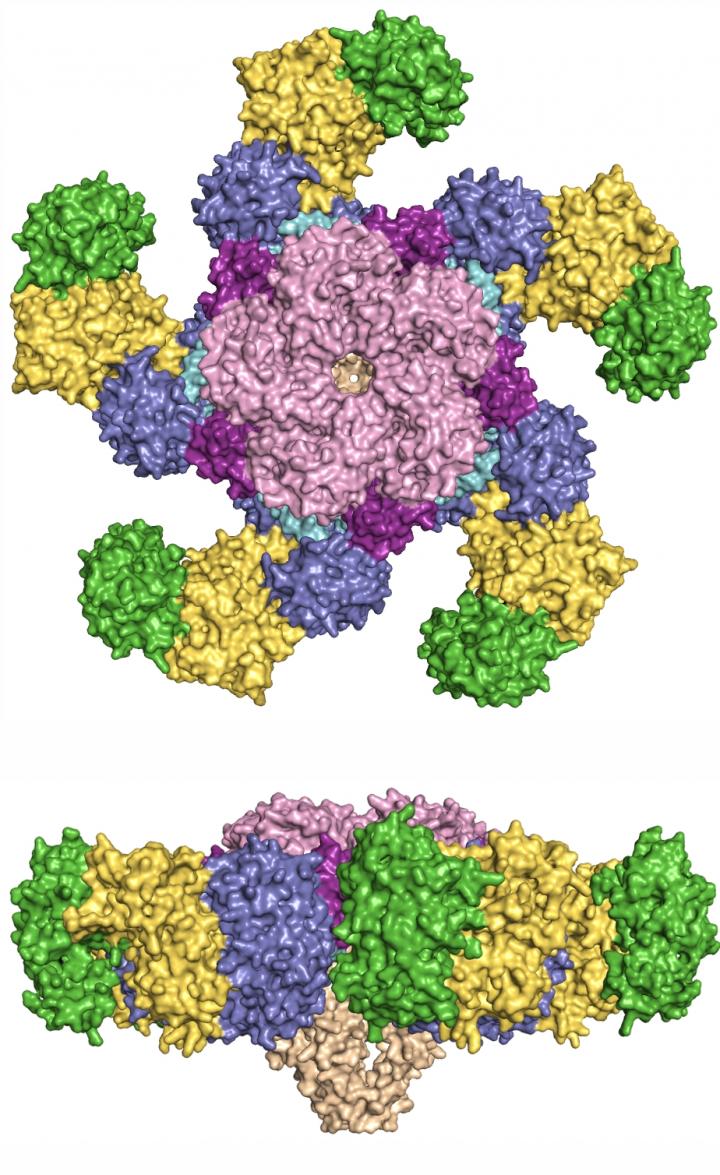
Using cryo-electron microscopy, Chai and co-authors observed that in the absence of bacterial effectors, ZAR1, together with the plant protein RKS1, is maintained in a latent state through interactions involving multiple domains of the ZAR1 protein. Upon infection, a bacterial effector modifies the plant ‘guardee’ PBL2, which then activates RKS1 resulting in huge conformational changes that first allow plants to swap ADP for ATP and then result in the assembly of a pentameric, wheel-like structure that the authors term the ‘ZAR1 resistosome’.
One striking feature of this structure is its similarity with animal NLR proteins, which, once activated, also assemble into wheel-like structures that act as signaling platforms for cell death execution and immune signaling. However, one important difference between the structures offers a tantalizing clue as to how ZAR1 induces cell death. The authors could identify a highly ordered funnel-like structure in ZAR1 that tethers the resistosome to the plasma membrane and is required for cell death and disease resistance. The authors speculate that ZAR1 may form a pore in the plasma membrane and in this way perturb cellular function leading to immune signaling and cell death.
Other plant NLRs also assemble into complexes that associate with the plasma membrane and it is thus highly likely that Chai’s findings have important general implications for understanding plant immunity. MPIPZ director Paul Schulze-Lefert, who was not involved in the studies, is in no doubt about the importance of the new studies: “This will become textbook knowledge.”
###
Media Contact
Jijie Chai
[email protected]




